Two New Cytotoxic Indole Alkaloids from a Deep-Sea Sediment Derived Metagenomic Clone
Abstract
:1. Introduction
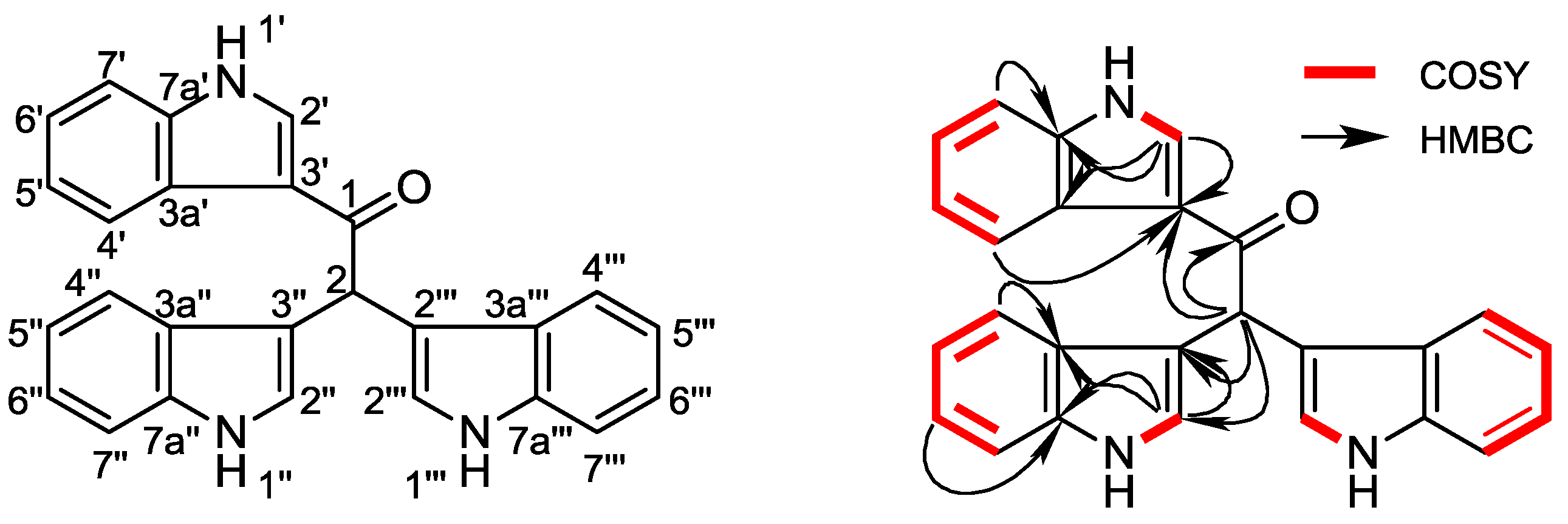
2. Results and Discussion
| Position | δH (J in Hz) | δC, Multiple | 1H-1H COSY | HMBC | |
|---|---|---|---|---|---|
| 1 | 193.9 | C | |||
| 2 | 6.41 s | 42.7 | CH | C-1, 3′, 3″, 3‴, 4″, 4‴ | |
| 1′ | 11.95 br.d (2.5) | H-2′ | n.o. a | ||
| 2′ | 8.74 d (3.2) | 133.8 | CH | H-1′ | C-3′, 3a′, 7a′ |
| 3′ | 126 | C | |||
| 3a′ | 115.6 | C | |||
| 4′ | 7.43 br.d (7.3) | 112 | CH | H-5′ | C-3′, 6′, 7a′ |
| 5′ | 7.13 td (7.1, 1.2) | 121.6 | CH | H-4′, H-6′ | C-3a′, 7′ |
| 6′ | 7.17 td (7.1, 1.2) | 122.7 | CH | H-5′, H-7′ | C4′, 7a′ |
| 7′ | 8.19 br.d (7.3) | 121.5 | CH | H-6′ | C-3a′, 6′, 7a′ |
| 7a′ | 136.6 | C | |||
| 1″, 1‴ | 10.86 br.d (1.7) | H-2″, 2‴ | |||
| 2″, 2‴ | 7.24 br.d (2.2) | 124 | CH | H-1″, 1‴ | C-3″, 3a″, 7a″, C-3‴, 3a‴, 7a‴ |
| 3″, 3‴ | 114.3 | C | |||
| 3a″, 3a‴ | 126.7 | C | |||
| 4″, 4‴ | 7.65 br.d (8.1) | 119.3 | CH | H-5″, 5‴ | C-3″, 6″, 7a″, C-3‴, 6‴, 7a‴ |
| 5″, 5‴ | 6.90 td (8.1, 1.0) | 118.2 | CH | H-4″, 4‴, H-6″, 6‴ | C-3a″, 7″, C-3a‴, 7‴ |
| 6″, 6‴ | 7.02 td (7.1, 1.0) | 120.8 | CH | H-5″, 5‴, H-7″, 7‴ | C4″, 7a″, C4‴, 7a‴ |
| 7″, 7‴ | 7.31 br.d (8.1) | 111.3 | CH | H-6″, 6‴ | C-3a″, 6″, 7a″, C-3a‴, 6‴, 7a‴ |
| 7a″, 7a‴ | 136.1 | C | |||

3. Experimental Section
3.1. General Experimental Procedures
3.2. Fermentation
3.3. Extraction and Isolation
| Position | δH (J in Hz) | δC, Multiple | 1H-1H COSY | HMBC | |
|---|---|---|---|---|---|
| 1 | 7.73 br.s | C-2, 3, 3a | |||
| 2 | 65.0 | C | |||
| 3 | 203.3 | C | |||
| 3a | 117.7 | C | |||
| 4 | 7.44 br.d (7.3) | 124.3 | CH | H-5 | C-6, 7a |
| 5 | 6.73 td (7.1, 0.5) | 117.1 | CH | H-4, H-6 | C-7 |
| 6 | 7.50 td (8.3, 1.3) | 137.5 | CH | H-5, H-7 | C-4, 7a |
| 7 | 6.90 br.d (8.3) | 111.9 | CH | H-6 | C-5 |
| 7a | 160.5 | C | |||
| 1′ | 11.03 br.s. | H-2′ | n.o. | ||
| 2′ | 7.37 d (2.4) | 123.5 | CH | H-1′ | C-2′, 3a′, 7a′ |
| 3′ | 114.4 | C | |||
| 3a′ | 124.8 | C | |||
| 4′ | 7.10 br.d (8.1) | 119.5 | CH | H-5′ | C-7a′ |
| 5′ | 6.80 td (8.1, 1.0) | 118.5 | CH | H-4′, H-6′ | C-3a′, 7′ |
| 6′ | 7.01 td (7.1, 1.0) | 121.0 | CH | H-5′, H-7′ | C-5′, 7a′ |
| 7′ | 7.32 br.d (8.1) | 111.6 | CH | H-6′ | C-3a′ |
| 7a′ | 136.6 | C | |||
| 2-CH3 | 1.62 s | 38.0 | CH | C-2, 3, 3′ | |
3.4. Cytotoxic Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Deming, J.W. Deep ocean environmental biotechnology. Curr. Opin. Microbiol. 1998, 9, 283–287. [Google Scholar]
- Lim, H.K.; Chung, E.J.; Kim, J.C.; Choi, G.J.; Jang, K.S.; Chung, Y.R.; Cho, K.Y.; Lee, S.W. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 7768–7777. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- Banik, J.J.; Brady, S. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr. Opin. Microbiol. 2010, 13, 603–609. [Google Scholar] [CrossRef]
- Chen, L.; Tang, X.X.; Zheng, M.; Yi, Z.W.; Xiao, X.; Qiu, Y.K.; Wu, Z. A novel indole alkaloid from deep-sea sediment metagenomic clone-derived Escherichia coli fermentation broth. J. Asian Nat. Prod. Res. 2011, 13, 444–448. [Google Scholar] [CrossRef]
- Gillespie, D.E.; Brady, S.F; Bettermann, A.D.; Cianciotto, N.P. Isolation of antibiotics turbomycins A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microb. 2002, 68, 4301–4306. [Google Scholar]
- Wu, Z.; Xiao, X.; Qiu, Y.K.; Tang, X.X.; Chen, L.; Yi, Z.W.; Zhao, H.Y.; Zhao, M. A Tri-Indole Compound, Preparation Method and Application Thereof. CN 102070507 A, 25 May 2011. [Google Scholar]
- Abe, T.; Kukita, A.; Akiyama, K. Isolation and structure of a novel biindole pigment substituted with an ethyl group from a metagenomic library derived from the marine sponge Halichondria okadai. Chem. Lett. 2012, 41, 728–729. [Google Scholar] [CrossRef]
- Zhuang, P.; Tang, X.X.; Yi, Z.W.; Qiu, Y.K.; Wu, Z. Two new compounds from marinederived fungus Penicillium sp. F11. J. Asian Nat. Prod. Res. 2012, 14, 197–203. [Google Scholar] [CrossRef]
- Brady, S.F.; Clardy, J. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew. Chem. Int. Ed. Engl. 2005, 44, 7063–7065. [Google Scholar] [CrossRef]
- Craig, J.W.; Chang, F.Y.; Brady, S.F. Natural products from environmental DNA hosted in Ralstonia metallidurans. ACS Chem. Biol. 2009, 4, 23–28. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yan, X.; Tang, X.-X.; Chen, L.; Yi, Z.-W.; Fang, M.-J.; Wu, Z.; Qiu, Y.-K. Two New Cytotoxic Indole Alkaloids from a Deep-Sea Sediment Derived Metagenomic Clone. Mar. Drugs 2014, 12, 2156-2163. https://doi.org/10.3390/md12042156
Yan X, Tang X-X, Chen L, Yi Z-W, Fang M-J, Wu Z, Qiu Y-K. Two New Cytotoxic Indole Alkaloids from a Deep-Sea Sediment Derived Metagenomic Clone. Marine Drugs. 2014; 12(4):2156-2163. https://doi.org/10.3390/md12042156
Chicago/Turabian StyleYan, Xia, Xi-Xiang Tang, Lin Chen, Zhi-Wei Yi, Mei-Juan Fang, Zhen Wu, and Ying-Kun Qiu. 2014. "Two New Cytotoxic Indole Alkaloids from a Deep-Sea Sediment Derived Metagenomic Clone" Marine Drugs 12, no. 4: 2156-2163. https://doi.org/10.3390/md12042156





