1. Introduction
Currently, about 60% of anticancer drugs originate from natural products, their derivatives, or mimics [
1]. Researchers have recognized that the incidence of biological activity in marine compounds is higher than in terrestrial-derived molecules [
2,
3]. The National Cancer Institute, USA, preclinical cytotoxicity screening demonstrated that approximately 1% of the tested marine samples showed antitumor potential compared to 0.1% of the tested terrestrial samples [
4]. This 10-fold higher hit rate is linked to the greater chemical diversity associated with marine organisms, which is a product of the very different conditions found in the oceans, such as high salinity, high pressure, and relatively constant temperature [
5]. Furthermore, it is thought that the immense competition for food and space challenges marine organisms to produce secondary metabolites for protection and survival [
6].
Marine-derived compounds encompass a wide range of chemical classes, including terpenes, shikimates, polyketides, acetogenins, peptides, alkaloids, and many others [
5]. It is estimated that there is US$563 billion–$5.69 trillion worth of anticancer marine-derived drugs still to be discovered [
7]. Furthermore, only a small percentage of the world’s marine biodiversity has been examined for biodiscovery, and it has been predicted that 90.4%–92.6% of marine compounds still remain undiscovered [
7]. Many marine compounds have generated interest for their cytotoxicity against multiple tumor types. The first marine-derived anticancer drug was ara-C (Cytosar-U
®), inspired by the unusual nucleosides spongothymidine and spongouridine isolated from the sponge
Cryptotethya crypta in the 1950s [
8]. Among the marine invertebrates, ascidians have been a plentiful source of cytotoxic compounds. Analysis of the first six marine-derived drugs that have made anticancer clinical trials showed that three were isolated from ascidians [
3]. The ascidian-derived compounds that have made clinical trials as antitumor agents are didemnin B [
9], ecteinascidin 743 [
10,
11], and aplidine [
12].
Breast cancer is the most common cancer in woman from developed countries [
13]. For American women the chance of developing this type of cancer during a lifetime is about 12.4%, being 1.8% for women aged between 20–34 years, and 22.2% for women that are 45–54 years old [
13]. It is also a major health problem for Australian woman, since it is the most common non-skin cancer, representing 28% of all reported cancers in females, and the second highest cause of cancer-related death in females [
14]. Chemotherapeutics are usually used to treat patients in stage 2 or later stages of the disease, which have a higher risk of recurrence [
15]. Different chemotherapeutics (anthracyclines, taxanes, alkylating agents, antimetabolites,
etc.) alone or in combination are the main treatment for estrogen receptor (ER) negative (ER-negative) patients but are also used in combination with hormone blocking therapy for ER-positive patients [
16].
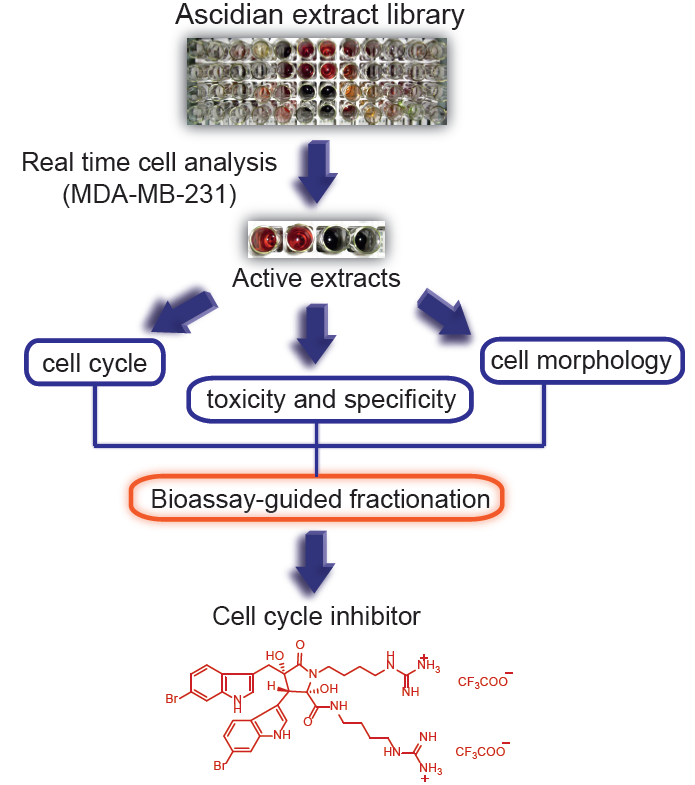
In order to isolate new cytotoxic compounds with potentially new mechanisms of action in breast cancer, we screened 143 ascidian extracts in MDA-MB-231 cells (ER-negative) using a real time cell analyzer (RTCA) [
17]. In addition, we conducted biological evaluation of the active extracts for effects on cell number, cell cycle distribution and cell morphology in order to prioritize the extracts for bioassay-guided fractionation. Following this approach, we isolated the previously reported natural product, eusynstyelamide B, which has never had cytotoxicity data reported.
3. Experimental Section
3.1. General
NMR spectra were recorded at 30 °C on a Varian 600 MHz spectrometer equipped with a triple resonance cold probe. The 1H and 13C chemical shifts were referenced to the solvent peak for DMSO-d6 at δH 2.49 and δC 39.5, respectively. LRESIMS were recorded on a Applied Biosystems Mariner Biospectrometry TOF workstation using either negative or positive electrospray ionization, and a mobile phase of 1:1 MeOH:H2O. The [α]D was measured on a JASCO P-1020 polarimeter The CD spectrum was recorded on a JASCO J-715 spectopolarimeter. Phenomenex SPE polypropylene cartridges (10 × 50 mm) with a 20 µm nylon frit inserted were used for small-scale extractions of the 143 ascidian samples. A Bio-line orbital shaker was used for the large-scale ascidian extraction. Alltech Davisil 30–40 µm 60 Å C18 bonded silica was used to pre-adsorb the ascidian extract prior to HPLC separation. An Alltech stainless steel guard cartridge (10 × 30 mm) was used for packing the pre-adsorbed material. A Waters 600 pump equipped with a Waters 996 PDA detector and a Waters 717 autosampler were used for HPLC separations. LC/MS analysis was performed on a Waters Alliance system (2790) using a Phenomenex Luna C18 3 µm 100 Å column (4.6 × 50 mm). A C18 Betasil 5 µm 143 Å column (21.2 mm × 150 mm) was used for semi-preparative HPLC separation. All solvents used for chromatography, [α]D CD, and MS were Lab-Scan HPLC grade, and the H2O was Millipore Milli-Q PF filtered.
3.2. Ascidian Material
A total of 143 ascidian specimens (Nature Bank Biota Library (
http://www.nature-bank.com.au)) belonging to the family Didemnidae were selected for the generation of the extract library. The selection was based on the amount of material available and exclusion of species that have been extensively studied [
22]. All marine samples were collected from the Great Barrier Reef (Queensland, Australia) and taxonomically identified by the Queensland Museum. Samples were kept frozen prior to freeze-drying and extraction. Voucher samples have been lodged at the Queensland Museum, Brisbane, Australia.
3.4. Cell Culture
MDA-MB-231 (ATCC) and NFF (Dusan Zencak, Eskitis Institute) cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% (v/v) heat-inactivated FBS (Invitrogen, Carlsbad, CA, USA) at 37 °C in a 5% CO2 humidified environment.
3.7. Cell Morphology Analysis by Phase-Contrast Microscopy
Each well of a 6-well plate was seeded with 0.3 × 106 cells. After 24 h, the cells were treated as described above and incubated for 24 h. Alternatively, cells were treated with 2.5 µM EB or 0.1% DMSO for 96 h. The cells were then photographed using an objective with 20× magnification (Olympus IX70, Olympus, Tokyo, Japan).
3.9. Large-Scale Extraction of Didemnum candidum (Ascidian Code: 114)
The freeze-dried and ground raw material (10 g) of Didemnum candidum (ascidian code: 114) was poured into a separate conical flask (1 L); n-hexane (250 mL) was added and the flask was shaken at 200 rpm for 2 h. The n-hexane extract was filtered under gravity, and discarded. CH2Cl2:MeOH (4:1, 250 mL) was then added to the de-fatted ascidian sample in the conical flask and shaken at 200 rpm for 2 h. The resulting extract was filtered under gravity, and set aside. MeOH (250 mL) was added to the ascidian sample, and the flask shaken at 200 rpm for 2 h before filtration. Another volume of MeOH (250 mL) was then added and the MeOH/ascidian mixture was shaken for a further 16 h at 200 rpm, followed by gravity filtration. All CH2Cl2/MeOH extracts were combined and dried under reduced pressure to afford a crude extract (536 mg) that was stored at 4 °C prior to bioassay-guided fractionation studies.
3.11. Determination of the IC50 of Eusynstyelamide B (1)
The IC50 of 1 was determined using the AlamarBlue® assay (Invitrogen, Carlsbad, CA, USA). MDA-MB-231 cells were seeded into a polystyrene black clear-bottomed 96-well plate at a density of 4000 cells/well. After 24 h, the cells were treated with different concentrations of 1 (0–100 µM) and incubated for 72 h. AlamarBlue® assay was performed according to the manufacturer’s instruction (Invitrogen, Carlsbad, CA, USA). Average values of triplicates were calculated after background correction. The IC50 was calculated by non-liner regression analysis with GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA).
3.12. Western Blotting Experiments Using Eusynstyelamide B (1)
MDA-MB-231 cells (0.1 × 106) were seeded in a 6 well/plate and treated with 5 µM of compound 1 for the indicated times. As positive controls, cells were treated with doxorubicin (1 µM, 48 h), etoposide (25 µM, 24 h), chloroquine (25 µM, 48 h), taxol (2 nM, 24 h), or nocodazole (83 nM, 24 h) which were purchased from Sigma Aldrich (Sigma-Aldrich, St. Louis, MO, USA). 0.1% DMSO was used as vehicle control. At the end of the treatment, cells were harvested and lysed with lysis buffer containing protease inhibitor cocktail (Roche, Basel, Switzerland). Protein concentration was determined with a bicinchoninic protein assay kit (BCA assay, Thermo Fisher Scientific, Waltham, MA, USA). Thirty micrograms of protein lysates per sample were loaded onto a NuPAGE® 4%–12% Bis-Tris Gel (Invitrogen, Carlsbad, CA, USA) and separated by electrophoresis for 90 min at 135 V. Proteins were transferred to a nitrocellulose membrane by wet transfer for 1 h at 100 V. Primary antibodies were used as recommended by the manufacturer; PARP antibody (#9542, Cell Signaling, Danvers, MA, USA), LC3B (#3868, Cell Signaling, Danvers, MA, USA) and beta-actin (04-1116, Merck Millipore, Billerica, MA, USA). Membranes were developed with the appropriate horseradish peroxidase-conjugated secondary antibody (GE Healthcare, Pittsburgh, PA, USA) and visualized with a chemiluminescence reaction system (Immobilon Western Chemiluminescent HRP Substrate, Merck Millipore, Billerica, MA, USA) and documented on a ChemiDoc XRS system (Bio-Rad, Hercules, CA, USA). Proteins were quantified using Image Lab™ software (Bio-Rad, Hercules, CA, USA), normalized according to the respective beta-actin levels, and were expressed as fold-change relative to the control treatment.
3.13. Annexin V Assay Using Eusynstyelamide B (1)
The assay was carried out according to the manufacturer’s instruction (BioVision, Milpitas, CA, USA). Briefly, MDA-MB-231 (0.1 × 106) cells were seeded in a 6 well/plate and treated with 5 µM EB or 0.1% DMSO. After 96 h, cells were harvested, washed twice with cold DPBS (Invitrogen) and counted. 1 × 105 cells were resuspended in 500 µL 1× binding buffer, containing 1 µL (0.15 mg/mL) of FITC-conjugated Annexin V (BioVision, Milpitas, CA, USA) and 1 µL of propidium iodide (PI, 1 mg/mL, Sigma-Aldrich, St. Louis, MO, USA) and incubated at room temperature for 20 min in the dark. After electronic compensation, fluorescence of 20,000 cells were measured in the PI and FITC channels by flow cytometry (FACSCanto, BD Biosciences, San Jose, CA, USA), and data were analyzed with FASCDiva software (BD Biosciences, San Jose, CA, USA). Double negative-stained cells were counted as viable cells, PI-negative/FITC-positive cells were counted as early apoptotic cells and double positive-stained cells were counted as late apoptotic/necrotic cells. Results were analyzed using one-way ANOVA (p < 0.05).
4. Discussion
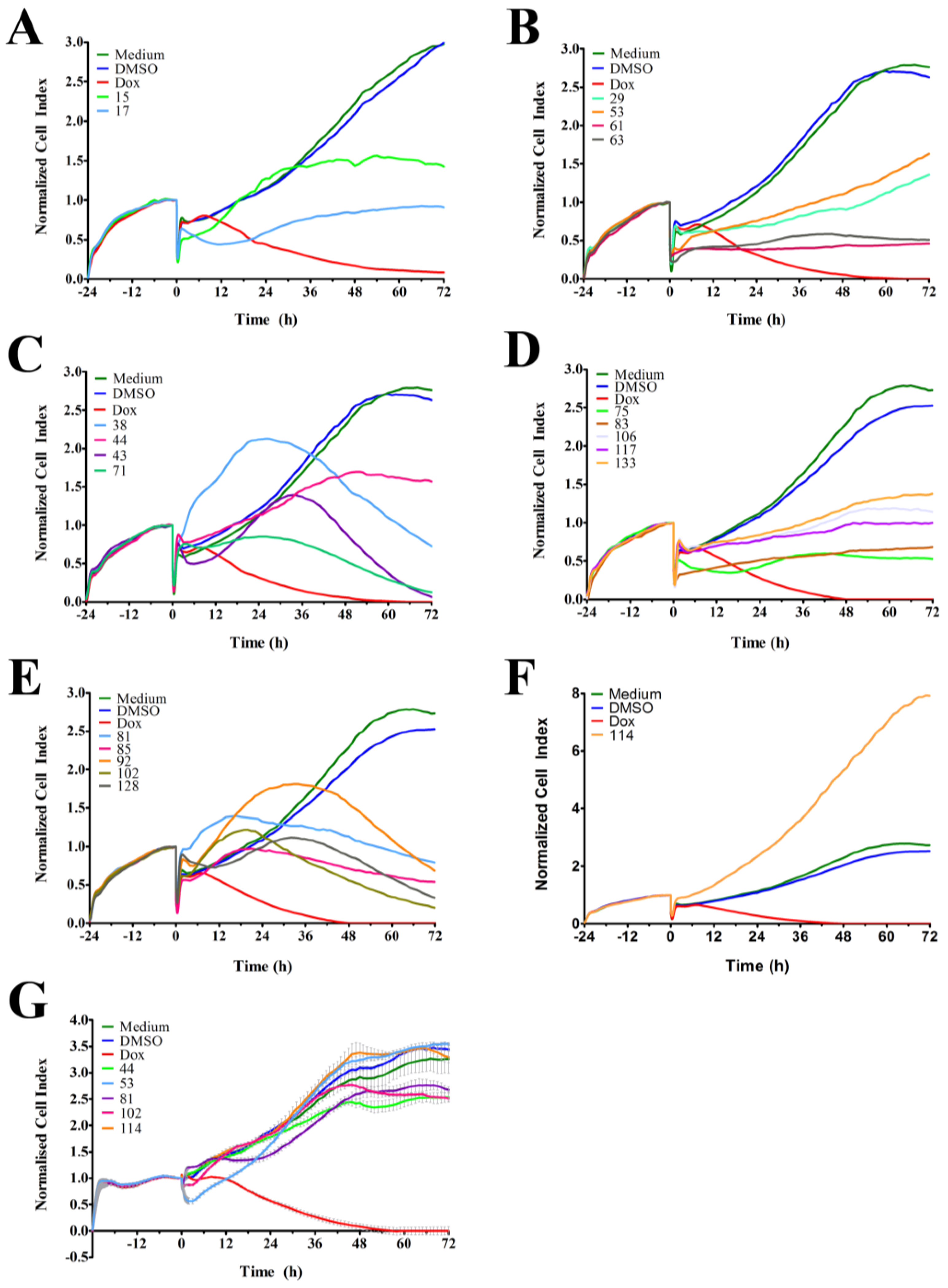
In order to identify and isolate new cytotoxic ascidian-derived compounds, 143 ascidian extracts were generated then subsequently screened in MDA-MB-231 breast cancer cells. Out of 143 ascidian extracts, we identified 21 extracts that affected the growth of MDA-MB-231 cells when tested by RTCA (
Figure 1). We observed substantial differences in the TCRPs among the active extracts. A few extracts caused an effect immediately after cells were treated, and both positive and negative effects on the CI were observed. However, the majority of active extracts required several hours or days to cause a divergence of the CI when compared to the controls. This time-dependent effect could have been due to the necessity of metabolic activation of the compound, accumulation of the compound, compound membrane solubility/uptake across membranes, accumulation of toxic intermediates, or end-product starvation among other reasons [
23]. In addition to the identification of active extracts, the RTCA-based TCRPs provided important information, such as optimal treatment period for subsequent end-point validation assays. A previous study, which tested 2000 small molecule compounds by RTCA, showed that compounds with a similar mode of action produced a similar TCRP, thereby providing predictive mechanistic information [
18]. However, the interpretation of extract-based TCRPs for the purpose of mechanism-of-action predictions might be limited due to the complexity of the active ascidian extracts. We did however observed that the TCRPs of most of the 21 active ascidian extracts fell into two previously reported and defined categories (
Figure 1) [
18]. The first category included bell shaped TCRPs, which has been found to be associated with inhibitors interfering with DNA synthesis, transcription and translation, such as 5-fluorouracil, etoposide, and MG132 [
18]. The second category, constituted TCRPs that showed an initial decline of the CI followed by a recovery and increase of the CI, which is typical for mitotic inhibitors, such as paclitaxel, nocodazole and colchicine [
18].
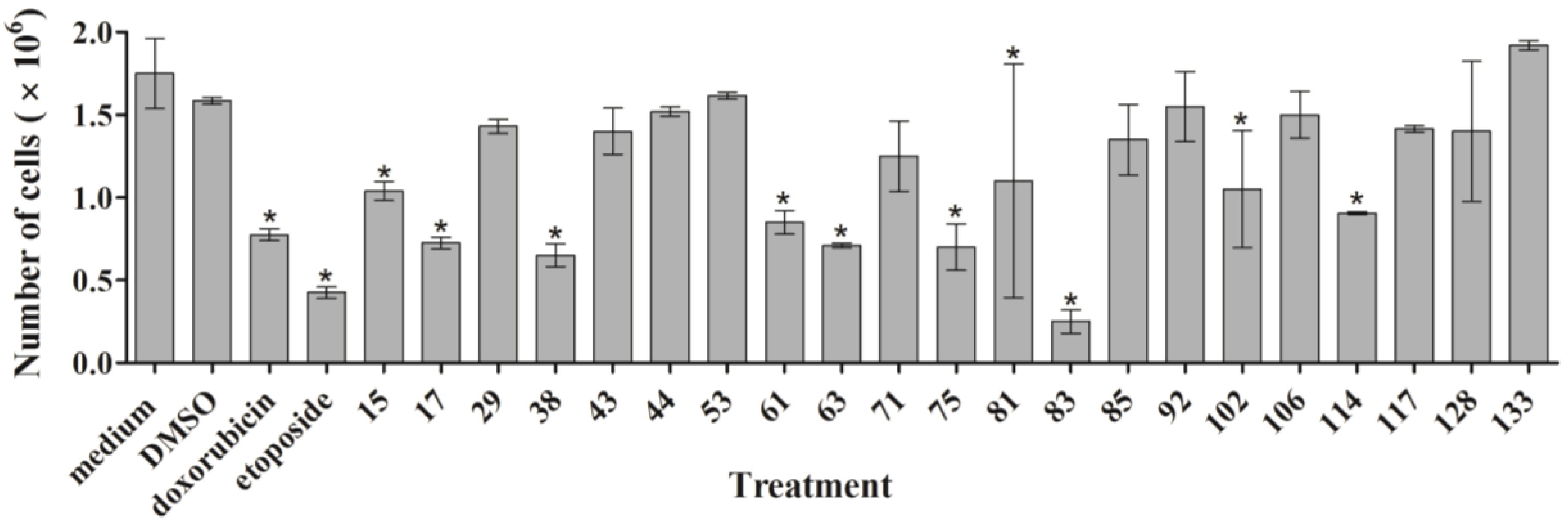
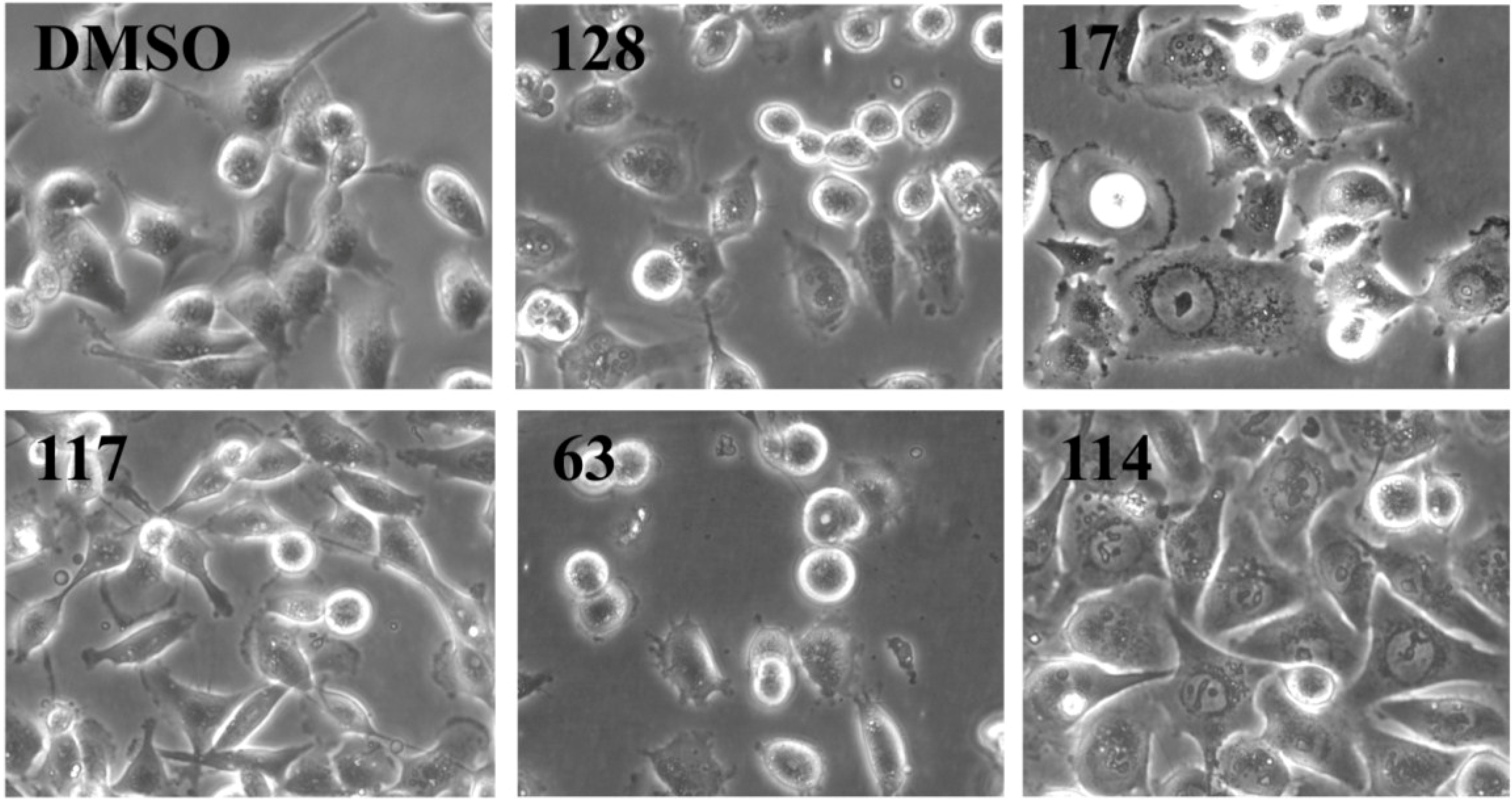
As outlined above, the RTCA measurement represents a composite figure of how a bioactive compound affects cell growth, morphology and adhesive characteristics of a cell culture. For validation purposes, we sought to address two of these cellular properties (growth and morphology) individually by performing cell count (
Figure 2) and phase contrast microscopy (
Figure 3,
Supplementary Figure S1) after treatment of MDA-MB-231 cells for 24 h. Cell counting demonstrated that nine extracts (15, 17, 38, 61, 63, 75, 83, 102, and 114) significantly reduced the number of cells compared to the control (
Figure 2). The RTCA data suggested that a longer incubation period might have facilitated more extracts reaching significance in reducing the cell number. Notably, none of the extracts increased the cell number after 24 h of treatment as implied by the RTCA data of extracts 38, 81, 92, 102, and 114, suggesting that changes to cell morphology and/or adherence caused the elevated CI. This observation was best exemplified by extracts 38 and 114 and highlighted the sensitivity of RTCA to changes in these cellular characteristics. Indeed, microscopy suggested that these extracts improved the adherence of MDA-MB-231 cells (38, 92, and 102), with fewer rounded and semi-attached cells present, and/or increased the cell size (81 and 114) when compared to control (
Figure 3,
Supplementary Figure S1). Furthermore, it confirmed the reduced cell numbers caused by extracts 15 and 17 (
Figure 2) by showing the typical signs of apoptosis,
i.e., loss of cell-cell contacts, cell shrinkage and membrane blebbing (
Figure 3). In summary, cell counting and microscopy were two relative easy and efficient assays that helped to de-convolute and interpret the complex RTCA data and facilitated the prioritization of extracts for bioassay-guided fractionation. Additional functional characterization of the extracts by cell cycle analysis of MDA-MB-231 cells (
Figure 4,
Supplementary Table S1) revealed that the majority of extracts caused arrest or delay of S phase (DNA synthesis) and G2/M phase (G2/mitosis), with extracts 15, 63, 81, and 114 leading to significant DNA fragmentation and cell death (subG1), possibly by inducing apoptosis. The absence of a significant subG1 cell population after treatment with extract 83, which caused the highest reduction in cell number among the extracts and a strong and immediate negative impact on the CI in RTCA suggested that pathways other than apoptosis or anoikis (e.g., autophagy) induced cell death. Interestingly, none of the extracts induced a G0/G1 cell cycle arrest. Consistent with our cell cycle results, previous studies have shown that ascidians are a rich source of DNA intercalating compounds and topoisomerase inhibitors [
24,
25]. Both classes of compounds cause DNA damage which can lead to S phase or G2/M arrest and are usually cytotoxic to different cell lines. RTCA screening of the 21 extracts in the non-malignant fibroblast cell line NFF revealed a relatively high level of general toxicity among the extracts, reinforcing the hypothesis of the presence of the aforementioned classes of compounds in the ascidian extracts. Only extracts 44, 53, 81, 102 and 114 were less toxic or cytostatic at the same dose in NFF cells when compared to the breast cancer cell line (
Figure 1G), suggesting some anti-cancer specificity.
The following criteria were used to prioritize the ascidian extracts for bioassay-guided fractionation: cell cycle effects, specific and general cytotoxicity, cell morphology, and logistic reasons, such as the amount of ascidian material available in Nature Bank (
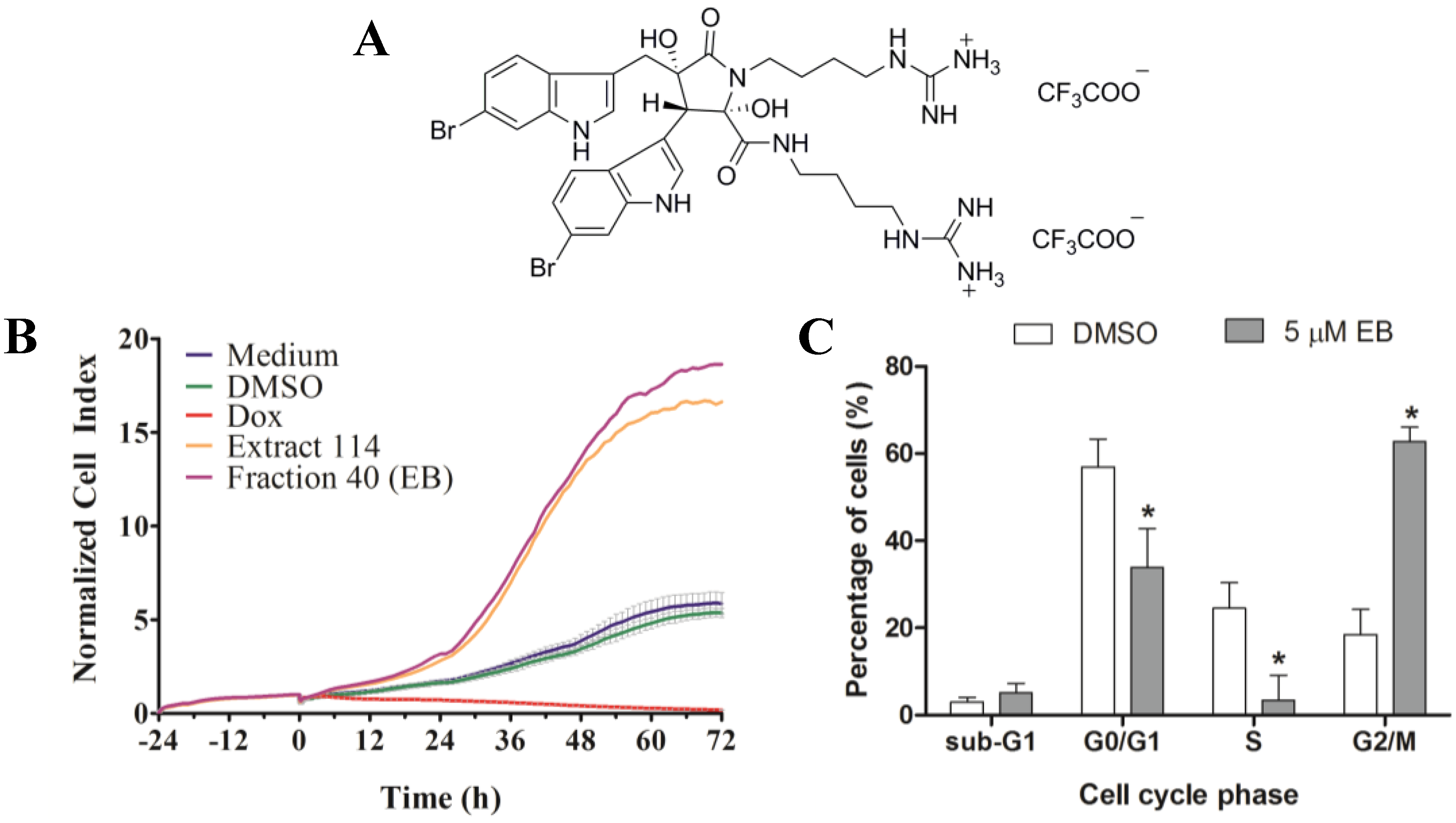
http://www.nature-bank.com.au). Only two extracts, 102 and 114, reduced the number of cells after treatment, caused cell cycle arrest, altered breast cancer cell morphology, and did not display cytotoxicity in NFF cells. Because of the larger amount of ascidian material available in NatureBank, extract 114 was selected for bioassay-guided fractionation, and yielded the known alkaloid dimer, eusynstyelamide B (EB,
1,
Supplementary Figures S3–S7). Compound
1 was previously isolated from the Great Barrier Reef ascidian
Eusynstyela latericus [
20]. It is the stereoisomer of eusynstyelamide A, which was first isolated from the ascidian
E. misakiensis, but whose chemical structure was initially misassigned [
20,
26]. Recently, the total synthesis of eusynstyelamide A was accomplished in six steps in 13% overall yield from 6-bromoindole, methyl glycidate, and Boc-protected agmatine [
27]. Motti
et al. suggested that
1 might be the result of the dimerization of two modified dipeptides formed from α-keto acid, derived from tryptophan, and agmatine by decarboxylation of arginine [
20]. Compound
1 has been shown to have mild antibacterial activity towards
Staphylococcus aureus (IC
50 6.5 µM), and inhibited the C4 plant regulatory enzyme pyruvate phosphate dikinase (IC
50 20 µM) [
20]. The enantiomer of
1, ent-eusynstyelamide B, isolated from the Arctic bryozoan
Tegella cf.
spitzbergensis was found to display anti-bacterial activity in
S. aureus (MIC 7.95 µM) and
Escherichia coli (MIC 15.9 µM) and was weakly active in
Candida albicans (MIC 127.2 µM) [
21]. The presence of stereoisomers and enantiomers of
1 in different phyla localized in different climatic zones suggests that the biosynthesis of this particular molecule may be performed by different microorganisms associated with the marine macro-organism [
28,
29]. Despite efforts by several research groups to determine the absolute configuration of the eusynstyelamide series, this currently remains unassigned. [
20,
21].
Previous work has found by colorimetric sulforhodamine B assay that
1 was not cytotoxic in the cancer cell lines MCF-7 (breast), SF-268 (CNS), and H-460 (lung) when tested up to 32 mM [
20]. Our discovery that
1 is cytotoxic in MDA-MB-231 cells, as shown by four different methods, might be due to differences in methodology and characteristics of the cell lines used. Our studies further revealed that
1 induced cell death of MDA-MB-231 cells through apoptosis. Interestingly,
1 has been reported to inhibit neural nitric oxide synthase (NOS1) with an IC
50 of 4.3 µM. NOS1 as well as its isoforms, inducible NOS (NOS2) and endothelial NOS (NOS3) are expressed in MDA-MB-231 cells [
30]. Thus, the nitric oxide synthase family could be a potential target of
1 in these cells.