Silaffins of Diatoms: From Applied Biotechnology to Biomedicine
Abstract
:1. Introduction
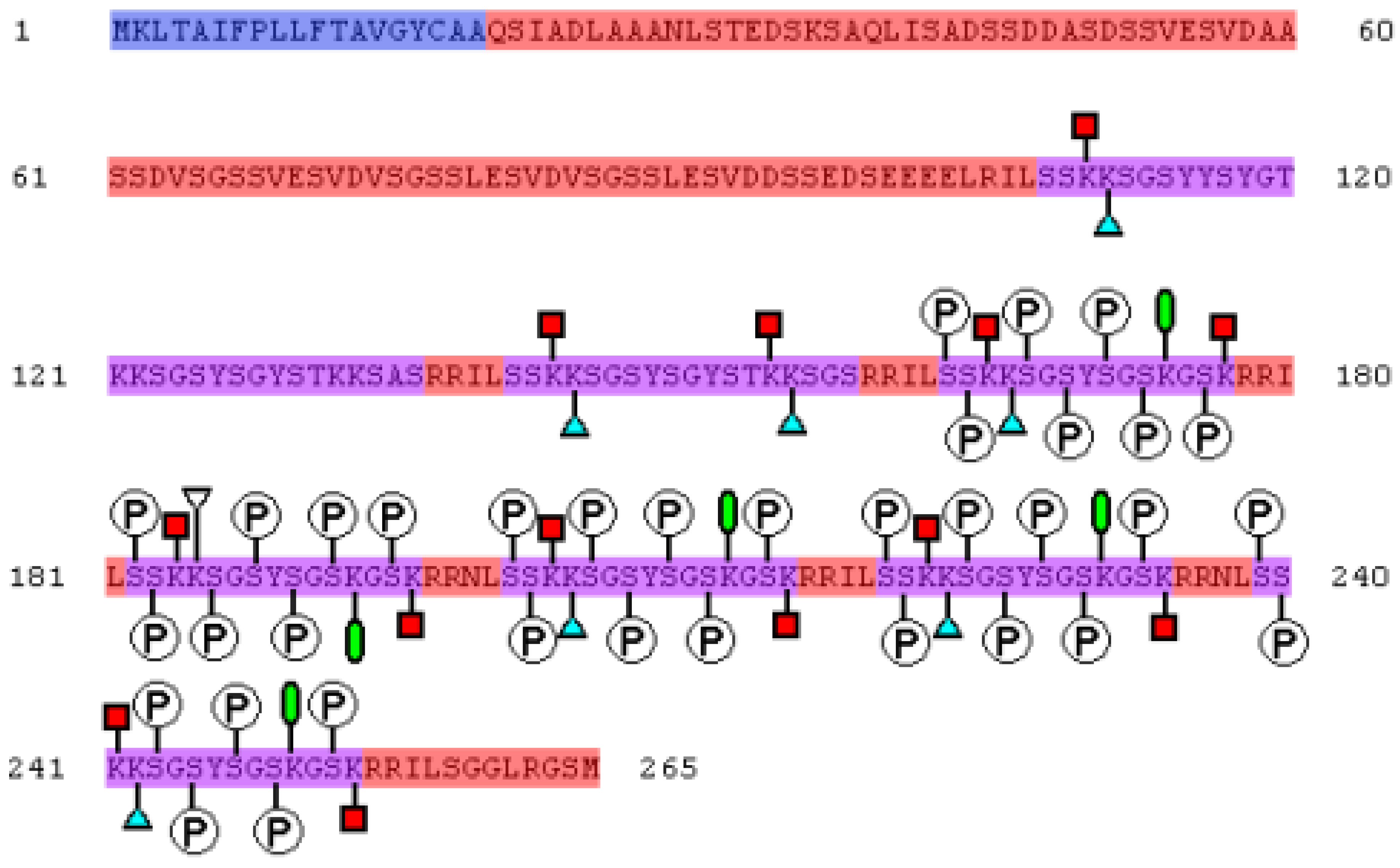
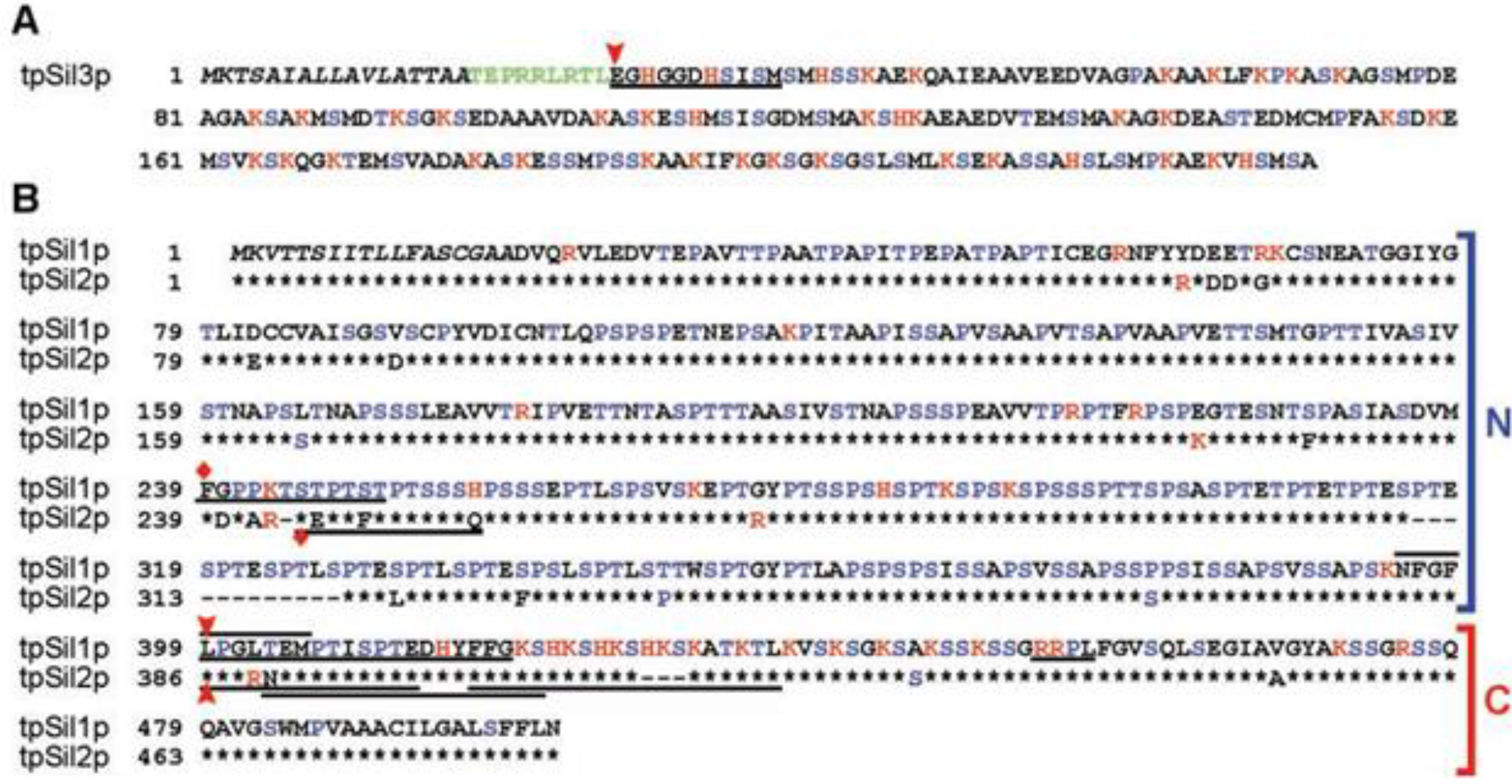
2. Mechanism of Biomineral Formation
2.1. Short Silaffins
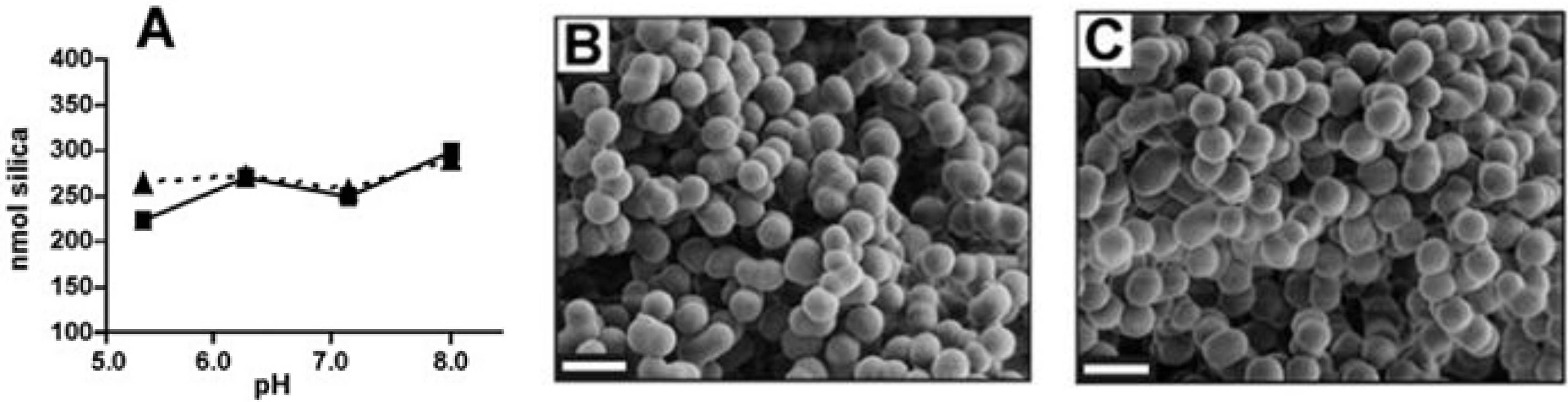


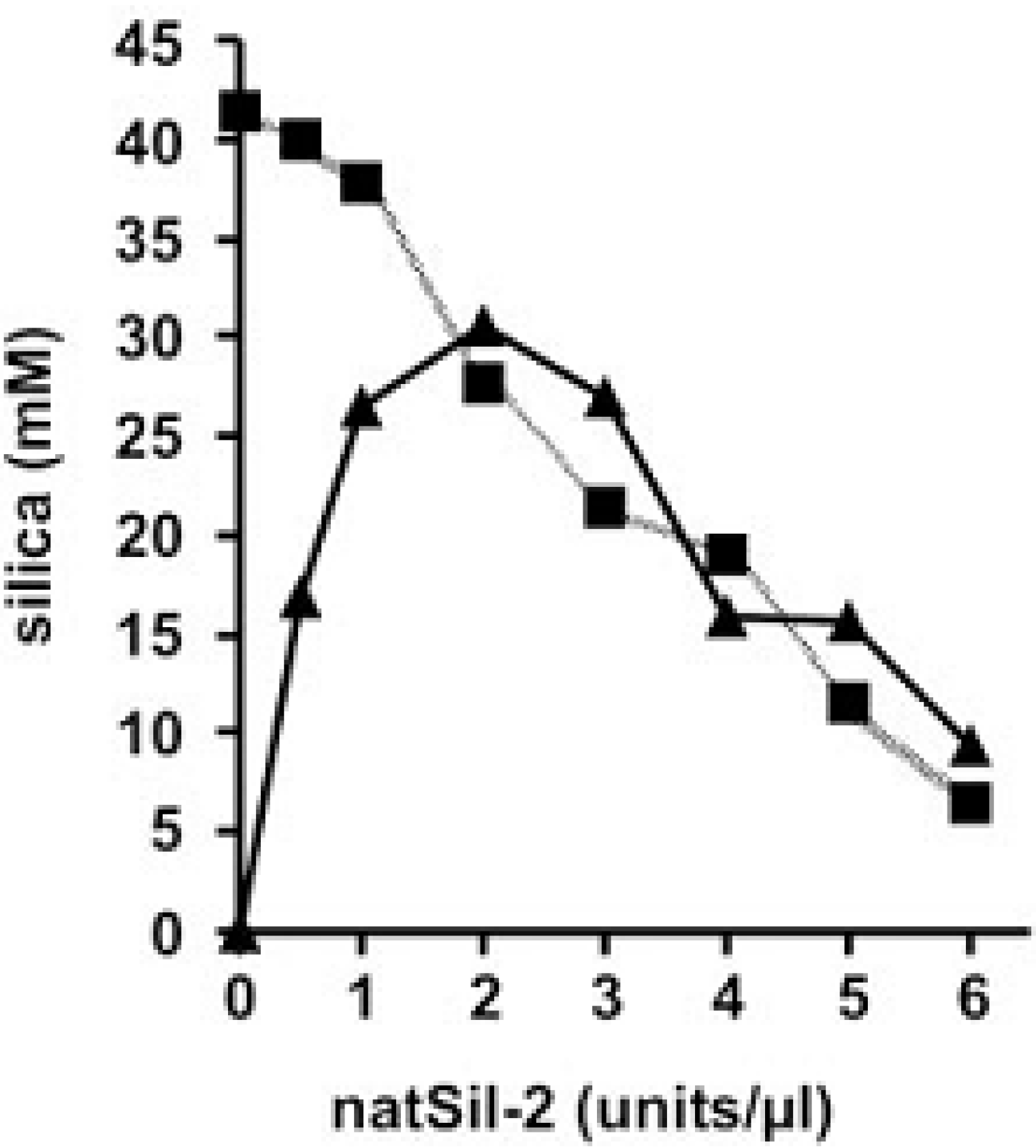
2.2. Long Silaffins
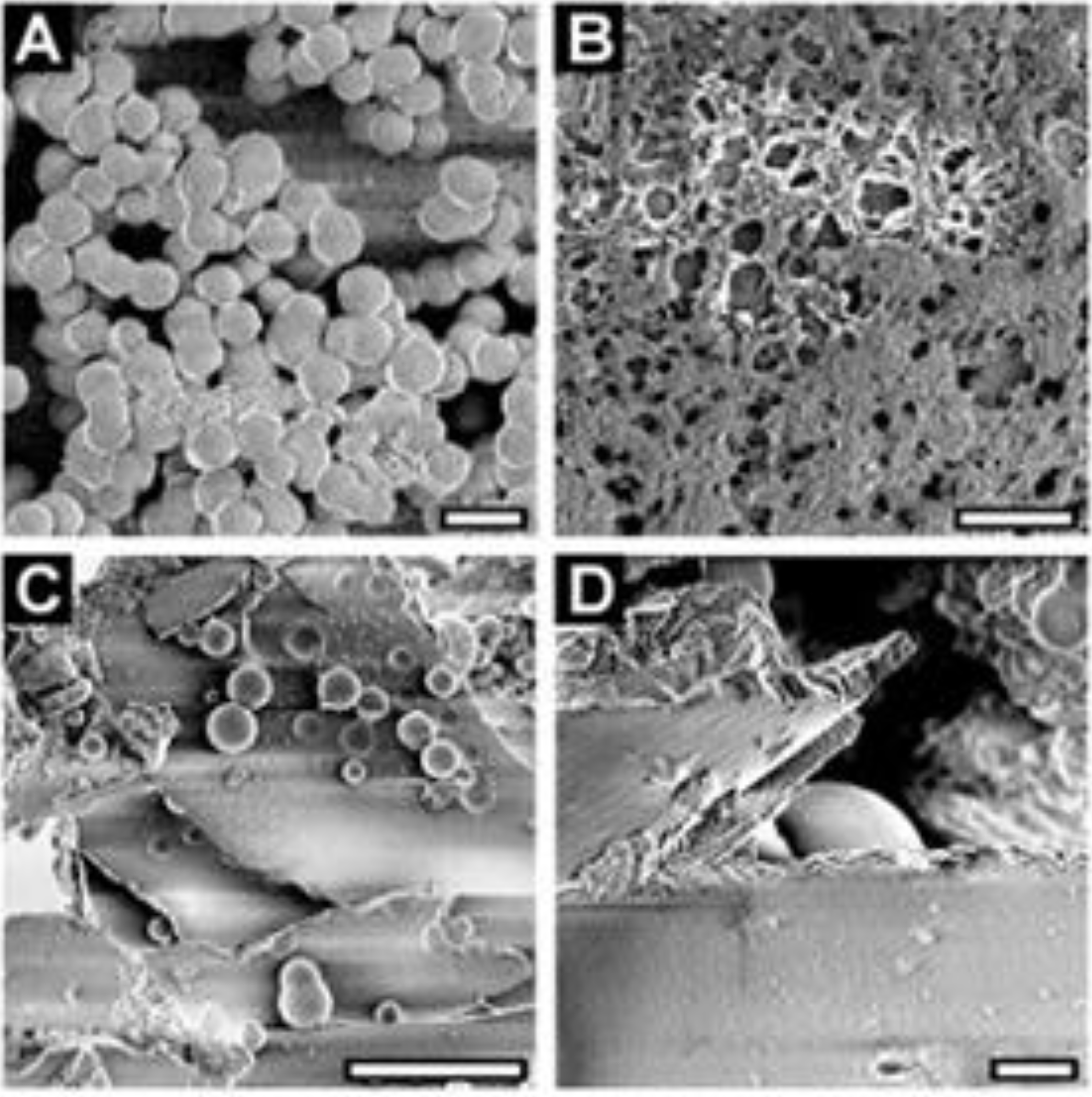
3. Practical Application of Silaffins
4. Conclusions
Acknowledgment
Conflicts of Interest
References
- Scala, S.; Bowler, C. Molecular insights into the novel aspects of diatom biology. Cell. Mol. Life Sci. 2001, 58, 1666–1673. [Google Scholar] [CrossRef]
- Kröger, N. Prescribing diatom morphology: Toward genetic engineering of biological nanomaterials. Curr. Op. Chem. Biol. 2007, 11, 662–669. [Google Scholar] [CrossRef]
- Poulsen, N.; Sumper, M.; Kröger, N. Biosilica formation in diatoms: Characterization of native silaffin-2 and its role in silica morphogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 12075–12080. [Google Scholar] [CrossRef]
- Sumper, M. A phase separation model for the nanopatterning of diatom biosilica. Science 2002, 295, 2430–2433. [Google Scholar] [CrossRef]
- Kröger, N.; Deutzmann, R.; Sumper, M. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 1999, 286, 1129–1132. [Google Scholar] [CrossRef]
- Kröger, N.; Poulsen, N. Diatoms—From cell wall biogenesis to nanotechnology. Annu. Rev. Genet. 2008, 42, 83–107. [Google Scholar] [CrossRef]
- Pamirsky, I.E.; Golokhvast, K.S. Search for homologues of proteins of primitive organisms biomineralization. Achiev. Life Sci. Russ. 2012, 4, 64–72. [Google Scholar]
- Golokhvast, K.S. Interaction of Organisms with Minerals; Far Eastern National Technical University: Vladivostok, Russia, 2010; pp. 1–115. [Google Scholar]
- Sumper, M.; Kröger, N. Silica formation in diatoms: The function of long-chain polyamines and silaffins. J. Mater. Chem. 2004, 14, 2059–2065. [Google Scholar] [CrossRef]
- Poulsen, N.; Kröger, N. Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana. J. Biol. Chem. 2004, 279, 42993–42999. [Google Scholar] [CrossRef]
- Sumper, M.; Hett, R.; Lehmann, G.; Wenzl, S. A code for lysine modifications of a silica biomineralizing silaffin protein. Angew. Chem. Int. Ed. 2007, 46, 8405–8408. [Google Scholar] [CrossRef]
- Wieneke, R.; Bernecker, A.; Riedel, R.; Sumper, M.; Steinem, C.; Geyer, A. Silica precipitation with synthetic silaffin peptides. Org. Biomol. Chem. 2011, 9, 5482–5486. [Google Scholar] [CrossRef]
- Gröger, C.; Lutz, K.; Brunner, E. Biomolecular self-assembly and its relevance in silica biomineralization. Cell. Biochem. Biophys. 2008, 50, 23–39. [Google Scholar] [CrossRef]
- Kröger, N.; Deutzmann, R.; Sumper, M. Silica-Precipitating peptides from diatoms. The chemical structure of silaffin-1a from Cylindrotheca fusiformis. J. Biol. Chem. 2001, 276, 26066–26070. [Google Scholar] [CrossRef]
- Kröger, N.; Lorenz, S.; Brunner, E.; Sumper, M. Self-Assembly of highly phosphorylated silaffins and their function in biosilica morphogenesis. Science 2002, 298, 584–586. [Google Scholar] [CrossRef]
- Patwardhan, S.V.; Shiba, K.; Schroder, H.C.; Muller, W.E.G.; Clarson, S.J.; Perry, C.C. The interaction of silicon with proteins: Part 2. The role of bioinspired peptide and recombinant proteins in silica polymerization. Sci. Technol. Silicones Silicone-Modif. Mat. 2007, 964, 328–347. [Google Scholar] [CrossRef]
- Whitlock, P.W.; Patwardhan, S.V.; Stone, M.O.; Clarson, S.J. Polymer Biocatalysis and Biomaterials II; Cheng, H.N., Gross, R.A., Eds.; Oxford University Press: Cary, NC, USA, 2008; pp. 412–433. [Google Scholar]
- Wong Po Foo, C.; Huang, J.; Kaplan, D.L. Lessons from Seashells: Silica mineralization via protein templating. Trends Biotechnol. 2004, 22, 577–585. [Google Scholar] [CrossRef]
- Patwardhan, S.V.; Clarson, S.J. Silicification and biosilicification. Part 4. Effect of template size on the formation of silica. J. Inorg. Organomet. Polym. 2002, 12, 109–116. [Google Scholar] [CrossRef]
- Kröger, N.; Deutzmann, R.; Bergsdorf, C.; Sumper, M. Species-Specific polyamines from diatoms control silica morphology. Proc. Natl. Acad. Sci. USA 2000, 97, 14133–14138. [Google Scholar] [CrossRef]
- Kröger, N.; Poulsen, N. Handbook of Biomineralization; Bäuerlein, E., Ed.; Weinheim Wiley-VCH: Weinheim, Baden-Württemberg, Germany, 2007; pp. 43–58. [Google Scholar]
- Sumper, M.; Brunner, E. Learning from diatoms: Nature’s tools for the production of nanostructured silica. Adv. Funct. Mater. 2006, 16, 17–26. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.; Schmid, A.M.M.; Edgar, L.A. Progress in Phycological Research; Round, F.E., Chapman, D.J., Eds.; Bristol Biopress: Bristol, UK, 1990; pp. 1–169. [Google Scholar]
- Van De Meene, A.M.L.; Pickett-Heaps, J.D. Valve morphogenesis in the centric diatom Proboscia alata Sundstrom. J. Phycol. 2002, 38, 351–363. [Google Scholar] [CrossRef]
- Tesson, B.; Hildebrand, M. Dynamics of silica cell wall morphogenesis in the diatom Cyclotella cryptica: Substructure formation and the role of microfilaments. J. Struct. Biol. 2010, 169, 62–74. [Google Scholar] [CrossRef]
- Robinson, D.H.; Sullivan, C.W. How do diatoms make silicon biominerals? Trends Biochem. Sci. 1987, 12, 151–154. [Google Scholar] [CrossRef]
- Wong Po Foo, C.; Patwardhan, S.V.; Belton, D.J.; Kitchel, B.; Anastasiades, D.; Huang, J.; Naik, R.R.; Perry, C.C.; Kaplan, D.L. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 259428–259433. [Google Scholar]
- Marner, W.D.; Shaikh, A.S.; Muller, S.J.; Keasling, J.D. Morphology of artificial silica matrices formed via autosilification of a silaffin/protein polymer chimera. Biomacromolecules 2008, 9, 1–5. [Google Scholar] [CrossRef]
- Mieszawska, A.J.; Nadkarni, L.D.; Perry, C.C.; Kaplan, D.L. Nanoscale control of silica particle formation via silk-silica fusion proteins for bone regeneration. Chem. Mater. 2010, 22, 5780–5785. [Google Scholar] [CrossRef]
- Okkyoung, C.; Byung-Chun, K.; Ji-Hye, A.; Kyoungseon, M.; Yong, H.K.; Youngsoon, U.; Min-Kyu, O.; Byoung-In, S. A biosensor based on the self-entrapment of glucose oxidase within biomimetic silica nanoparticles induced by a fusion enzyme. Enzym. Microb. Technol. 2011, 49, 441–445. [Google Scholar] [CrossRef]
- Brott, L.L.; Naik, R.R.; Pikas, D.J.; Kirkpatrick, S.M.; Tomlin, D.W.; Whitlock, P.W.; Clarson, S.J.; Stone, M.O. Ultrafast holographic nanopatterning of biocatalytically formed silica. Nature 2001, 413, 291–293. [Google Scholar] [CrossRef]
- Luckarift, H.R.; Spain, J.C.; Naik, R.R.; Stone, M.O. Enzyme immobilization in a biomimetic silica support. Nat. Biotechnol. 2004, 22, 211–213. [Google Scholar] [CrossRef]
- Luckarift, H.R.; Johnson, G.R.; Spain, J.C. Silica-Immobilized enzyme reactors; Application to cholinesterase-inhibition studies. J. Chromatogr. 2006, B843, 310–316. [Google Scholar]
- Nam, D.H.; Won, K.; Kim, Y.H.; Sang, B.I. A novel route for immobilization of proteins to silica particles incorporating silaffin domains. Biotechnol. Prog. 2009, 25, 1643–1649. [Google Scholar]
- Poulsen, N.; Berne, C.; Spain, J.; Kroger, N. Silica immobilization of an enzyme through genetic engineering of the diatom Thalassiosira pseudonana. Angew. Chem. Int. Ed. 2007, 46, 1843–1846. [Google Scholar] [CrossRef]
- Marner, W.D.; Shaikh, A.S.; Muller, S.J.; Keasling, J.D. Enzyme immobilization via silaffin-mediated autoencapsulation in a biosilica support. Biotechnol. Prog. 2009, 25, 417–423. [Google Scholar] [CrossRef]
- Kharlampieva, E.; Jung, C.M.; Kozlovskaya, V.; Tsukruk, V.V. Secondary structure of silaffin at interfaces and titania formation. J. Mat. Chem. 2010, 20, 5242–5250. [Google Scholar] [CrossRef]
- Kharlampieva, E.; Slocik, J.M.; Singamaneni, S.; Poulsen, N.; Kroger, N.; Naik, R.R.; Tsukruk, V.V. Protein-Enabled synthesis of monodisperse titania nanoparticles on and within polyelectrolyte matrices. Adv. Funct. Mat. 2009, 19, 2303–2311. [Google Scholar] [CrossRef]
- Sewell, S.L.; Wright, D.W. Biomimetic synthesis of titanium dioxide utilizing the R5 peptide derived from Cylindrotheca fusiformis. Chem. Mat. 2006, 18, 3108–3113. [Google Scholar] [CrossRef]
- Carvalho, R.N.; Burchardt, A.D.; Sena, F.; Mariani, G.; Mueller, A.; Bopp, S.K.; Umlauf, G.; Lettieri, T. Gene biomarkers in diatom Thalassiosira pseudonana exposed to polycyclic aromatic hydrocarbons from contaminated marine surface sediments. Aquat. Toxicol. 2011, 101, 244–253. [Google Scholar] [CrossRef]
- Nam, D.H.; Lee, J.-O.; Sang, B.-I.; Won, K.; Kim, Y.H. Silaffin peptides as a novel signal enhancer for gravimetric biosensors. Appl. Biochem. Biotechnol. 2013, 170, 25–31. [Google Scholar] [CrossRef]
- Annenkov, V.V.; Patwardhan, S.V.; Belton, D.; Danilovtseva, E.N.; Perry, C.C. A new stepwise synthesis of a family of propylamines derived from diatom silaffins and their activity in silicification. Chem. Commun. 2006, 14, 1521–1523. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pamirsky, I.E.; Golokhvast, K.S. Silaffins of Diatoms: From Applied Biotechnology to Biomedicine. Mar. Drugs 2013, 11, 3155-3167. https://doi.org/10.3390/md11093155
Pamirsky IE, Golokhvast KS. Silaffins of Diatoms: From Applied Biotechnology to Biomedicine. Marine Drugs. 2013; 11(9):3155-3167. https://doi.org/10.3390/md11093155
Chicago/Turabian StylePamirsky, Igor E., and Kirill S. Golokhvast. 2013. "Silaffins of Diatoms: From Applied Biotechnology to Biomedicine" Marine Drugs 11, no. 9: 3155-3167. https://doi.org/10.3390/md11093155



