The Relative Performance of a Benchtop Scanning Monochromator and Handheld Fourier Transform Near-Infrared Reflectance Spectrometer in Predicting Forage Nutritive Value
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Database
2.2. Laboratory Reference Methods
2.3. Instruments
2.4. Model Development
3. Results and Discussion
3.1. Sample Database
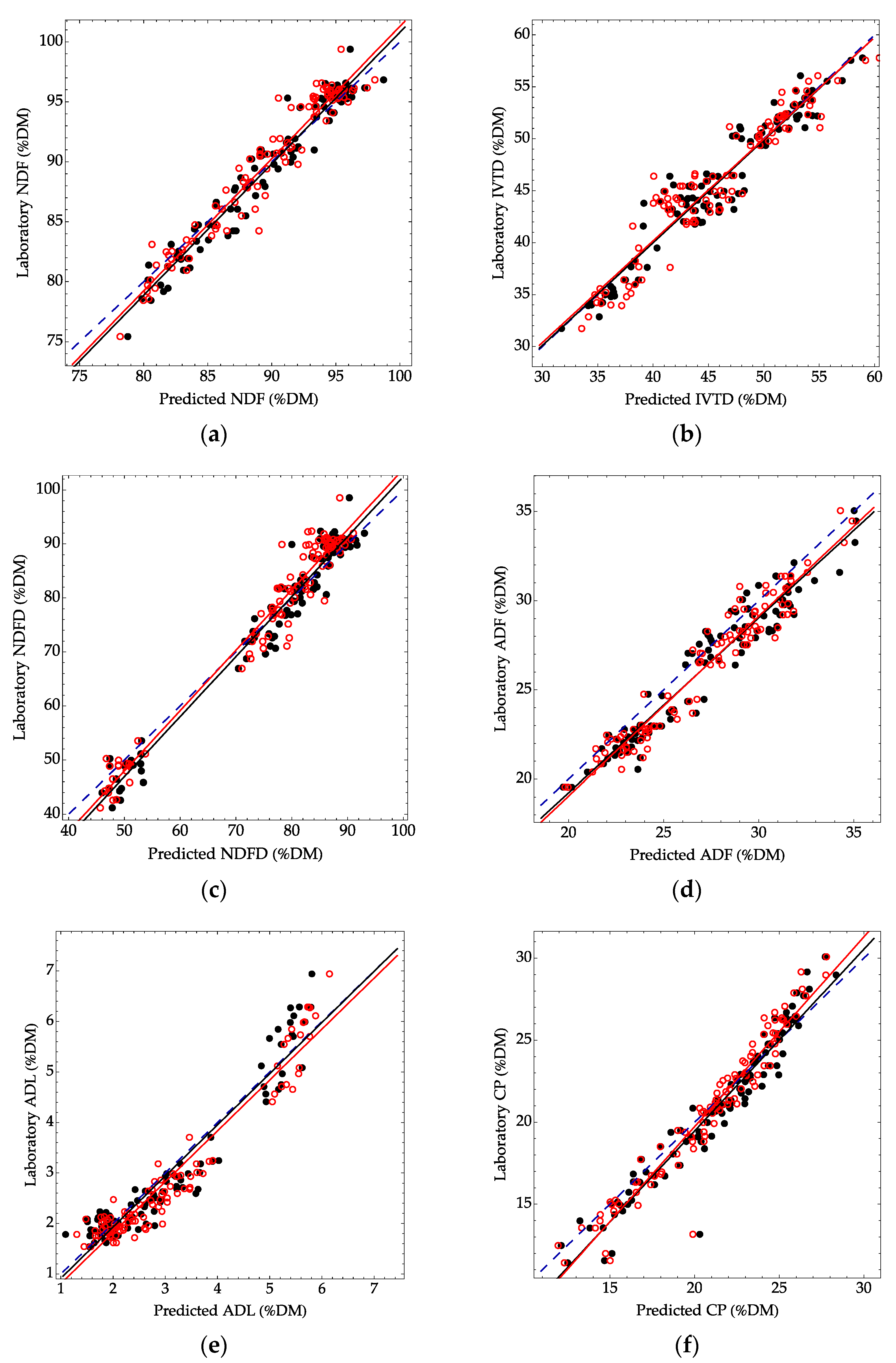
3.2. Relative Instrument Performance
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cherney, D.J.R.; Digman, M.F.; Cherney, J.H. Day-to-day variation in forage and mixed diets on commercial dairy farms in NY. Appl. Anim. Sci. 2021, 37, 11–20. [Google Scholar] [CrossRef]
- Sniffen, C.J.; Beverly, R.W.; Mooney, C.S.; Roe, M.B.; Skidmore, A.L.; Black, J.R. Nutrient requirements versus supply in the dairy cow: Strategies to account for variability. J. Dairy Sci. 1993, 76, 3160–3178. [Google Scholar] [CrossRef]
- Marten, G.C.; Halgerson, J.L.; Cherney, J.H. Quality prediction of small grain silage forages by near infrared reflectance spectroscopy. Crop Sci. 1983, 23, 94–96. [Google Scholar] [CrossRef]
- New NIR for Feed: Hard Work or a Smart Move? Available online: https://www.fossanalytics.com/en/news-articles/feed-and-forage/nir-instrument (accessed on 20 August 2021).
- Kormann, G.; Flohr, W. Ingredients measurement on self propelled forage harvesters. In Proceedings of the EuroAgEng, Sisole Bedford, UK, 2002. Abstract Number 02-PA-009. [Google Scholar]
- Wright, S.; Brumback, S.; Neibur, T.; Welle, R. Near Infrared Spectrometry for Real Time Analysis of Substances. U.S. Patent 6,483,583, 27 February 1997. [Google Scholar]
- Digman, M.F.; Shinners, K.J. Real-Time Moisture Measurement on a Forage Harvester Using Near-Infrared Reflectance Spectroscopy. Trans. ASABE 2008, 51, 1801–1810. [Google Scholar] [CrossRef]
- Cherney, D.J.R.; Digman, M.; Cherney, J.H. Handheld NIRS for forage evaluation. Comput. Electron. Agric. 2021, 190, 106469. [Google Scholar] [CrossRef]
- Acosta, J.J.; Castillo, M.S.; Hodge, G.R. Comparison of benchtop and handheld near-infrared spectroscopy devices to determine forage nutritive value. Crop Sci. 2020, 60, 3410–3422. [Google Scholar] [CrossRef]
- Berzaghi, P.; Cherney, J.H.; Casler, M. Prediction performance of portable near infrared reflectance instruments using preprocessed dried, ground forage samples. Comput. Electron. Agric. 2021, 182, 106013. [Google Scholar] [CrossRef]
- Rukundo, I.R.; Danao, M.G.; Mitchell, R.B.; Masterson, S.D.; Weller, C.L. Comparing the use of handheld and benchtop NIR spectrometers in predicting nutritional value of forage. Appl. Eng. Agric. 2021, 37, 171–181. [Google Scholar] [CrossRef]
- Mayr, S.; Beć, K.B.; Grabska, J.; Wiedemair, V.; Pürgy, V.; Popp, M.A.; Bonn, G.K.; Huck, C.W. Challenging handheld NIR spectrometers with moisture analysis in plant matrices: Performance of PLSR vs. GPR vs. ANN modelling. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 249, 119342. [Google Scholar] [CrossRef] [PubMed]
- Bec, K.B.; Grabska, J.; Siesler, H.W.; Huck, C.W. Handheld near-infrared spectrometers: Where are we heading? NIR News 2020, 31, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Valentine, M.E.; Karayilanli, E.; Cherney, J.H.; Cherney, D.J. Comparison of in vitro long digestion methods and digestion rates for diverse forages. Crop Sci. 2019, 59, 422–435. [Google Scholar] [CrossRef]
- Cunniff, P. (Ed.) Protein (crude) in animal feed. Combustion method (990.03). In Official Methods of Analysis of AOAC International, 15th ed.; AOAC: Arlington, VA, USA, 1995; Volume 3, pp. 154–196. [Google Scholar]
- Roberts, C.A.; Stuth, J.; Flinn, P. Analysis of Forages and Feedstuffs. In Near-Infrared Spectroscopy in Agriculture; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2015; pp. 229–267. [Google Scholar] [CrossRef]
- Van Roon, P.; Zakizadeh, J.; Chartier, S. Partial Least Squares tutorial for analyzing neuroimaging data. Quant. Methods Psychol. 2014, 10, 200–215. [Google Scholar] [CrossRef] [Green Version]
- Dardenne, P. Some Considerations about NIR Spectroscopy: Closing Speech at NIR-2009. NIR News 2010, 21, 8–14. [Google Scholar] [CrossRef]


| NDF | IVTD | NDFD | ADF | ADL | CP | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 284 | |||||||||||
| Units | %DM | |||||||||||
| SEL | 0.45 | 0.57 | 1.34 | 0.74 | 0.29 | 0.21 | ||||||
| Min. | 76 | 21 | 40 | 18 | 1.5 | 13 | ||||||
| Mean | 87 | 46 | 70 | 30 | 3.7 | 20 | ||||||
| Max. | 97 | 64 | 93 | 38 | 7.2 | 33 | ||||||
| Stdev. | 4.5 | 13 | 13 | 5.8 | 1.2 | 4.8 | ||||||
| FOSS | NEO | FOSS | NEO | FOSS | NEO | FOSS | NEO | FOSS | NEO | FOSS | NEO | |
| LVs | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| SEC | 1.4 | 1.5 | 2.3 | 2.4 | 3.3 | 3.5 | 1.5 | 1.5 | 0.44 | 0.44 | 1.3 | 1.3 |
| R2C | 0.91 | 0.89 | 0.97 | 0.97 | 0.94 | 0.93 | 0.93 | 0.93 | 0.87 | 0.87 | 0.93 | 0.93 |
| SECV | 1.5 | 1.6 | 2.4 | 2.4 | 3.6 | 3.7 | 1.6 | 1.7 | 0.47 | 0.46 | 1.3 | 1.3 |
| R2CV | 0.91 | 0.88 | 0.97 | 0.96 | 0.94 | 0.93 | 0.93 | 0.92 | 0.87 | 0.87 | 0.93 | 0.93 |
| NDF | IVTD | NDFD | ADF | ADL | CP | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 100 | |||||||||||
| Units | %DM | |||||||||||
| Min. | 76 | 32 | 41 | 20 | 1.5 | 11 | ||||||
| Mean | 87 | 46 | 77 | 26 | 2.9 | 21 | ||||||
| Max. | 95 | 58 | 99 | 35 | 6.9 | 30 | ||||||
| Stdev. | 4.2 | 6.5 | 16 | 3.8 | 1.4 | 4.4 | ||||||
| FOSS | NEO | FOSS | NEO | FOSS | NEO | FOSS | NEO | FOSS | NEO | FOSS | NEO | |
| R2P | 0.95 | 0.94 | 0.92 | 0.90 | 0.97 | 0.96 | 0.93 | 0.94 | 0.91 | 0.93 | 0.93 | 0.93 |
| RMSEP | 1.4 | 1.6 | 1.8 | 2.1 | 3.3 | 3.8 | 1.4 | 1.3 | 0.42 | 0.42 | 1.3 | 1.3 |
| SEP | 1.4 | 1.5 | 1.8 | 2.1 | 3.3 | 3.7 | 1.0 | 0.96 | 0.42 | 0.38 | 1.2 | 1.3 |
| Bias | −0.17 | −0.22 | −0.08 | −0.07 | −0.13 | −0.96 | −0.90 | 0.89 | −0.06 | 0.17 | −0.40 | 0.11 |
| Slope | 1.1 | 1.1 | 0.99 | 0.98 | 1.1 | 1.1 | 0.98 | 1.0 | 1.0 | 1.0 | 1.1 | 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Digman, M.F.; Cherney, J.H.; Cherney, D.J.R. The Relative Performance of a Benchtop Scanning Monochromator and Handheld Fourier Transform Near-Infrared Reflectance Spectrometer in Predicting Forage Nutritive Value. Sensors 2022, 22, 658. https://doi.org/10.3390/s22020658
Digman MF, Cherney JH, Cherney DJR. The Relative Performance of a Benchtop Scanning Monochromator and Handheld Fourier Transform Near-Infrared Reflectance Spectrometer in Predicting Forage Nutritive Value. Sensors. 2022; 22(2):658. https://doi.org/10.3390/s22020658
Chicago/Turabian StyleDigman, Matthew F., Jerry H. Cherney, and Debbie J. R. Cherney. 2022. "The Relative Performance of a Benchtop Scanning Monochromator and Handheld Fourier Transform Near-Infrared Reflectance Spectrometer in Predicting Forage Nutritive Value" Sensors 22, no. 2: 658. https://doi.org/10.3390/s22020658






