Advances in Analysis of Milk Proteases Activity at Surfaces and in a Volume by Acoustic Methods
Abstract
:1. Introduction
2. Milk Proteases
3. Surface Acoustic Methods
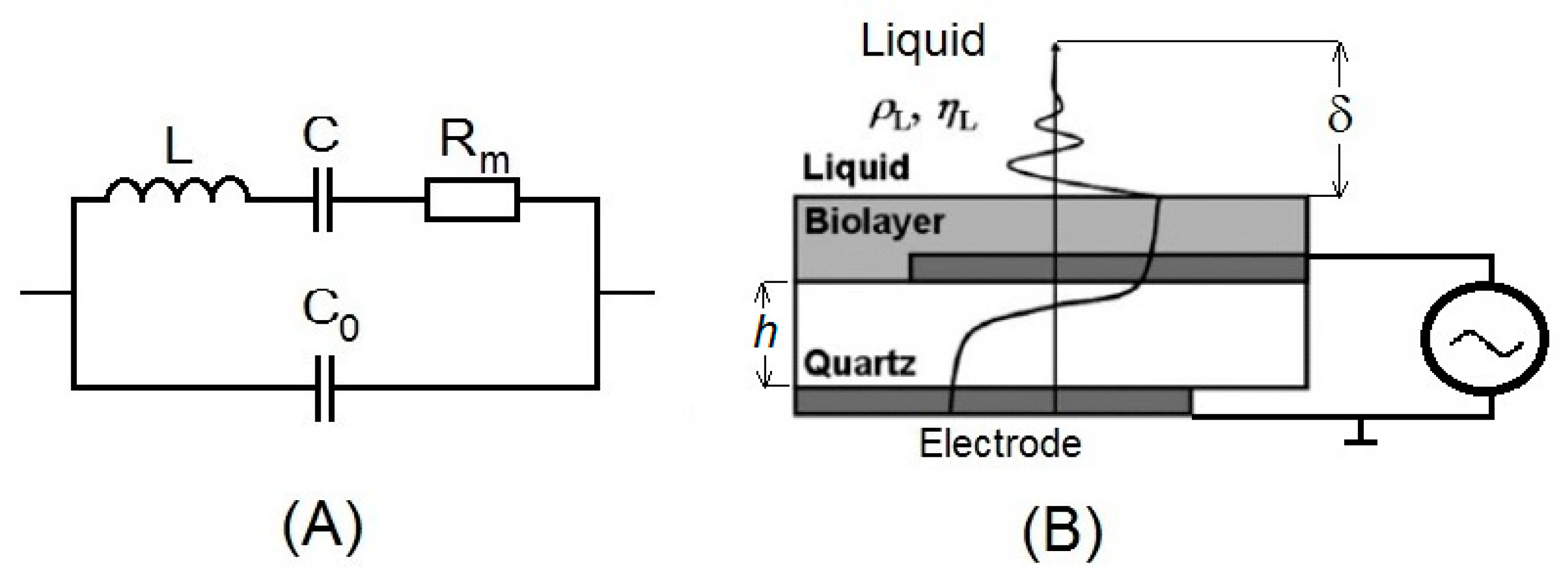
3.1. The Principles of QCM and TSM
3.2. The Principles of Electromagnetic Piezoelectric Sensors EMPAS
3.3. Immobilization of the Proteins at the Piezoelectric Transducers
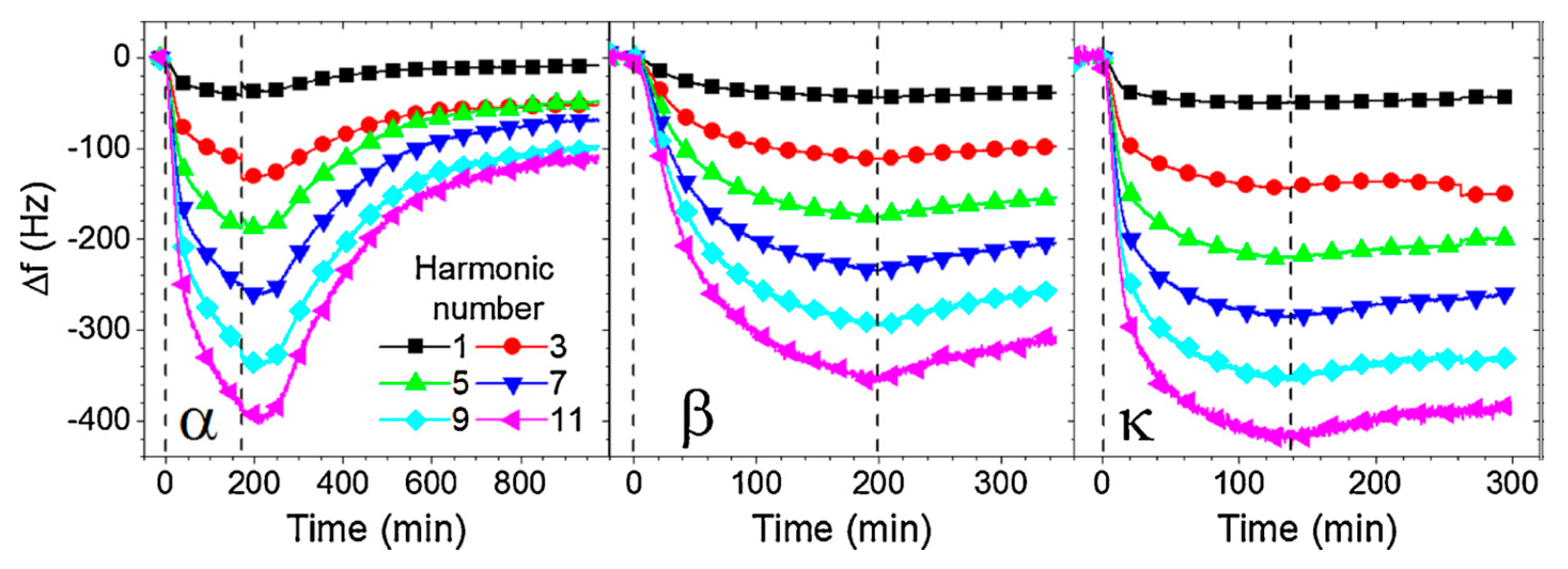
3.4. Application of Surface Acoustic Methods for Detection Proteases
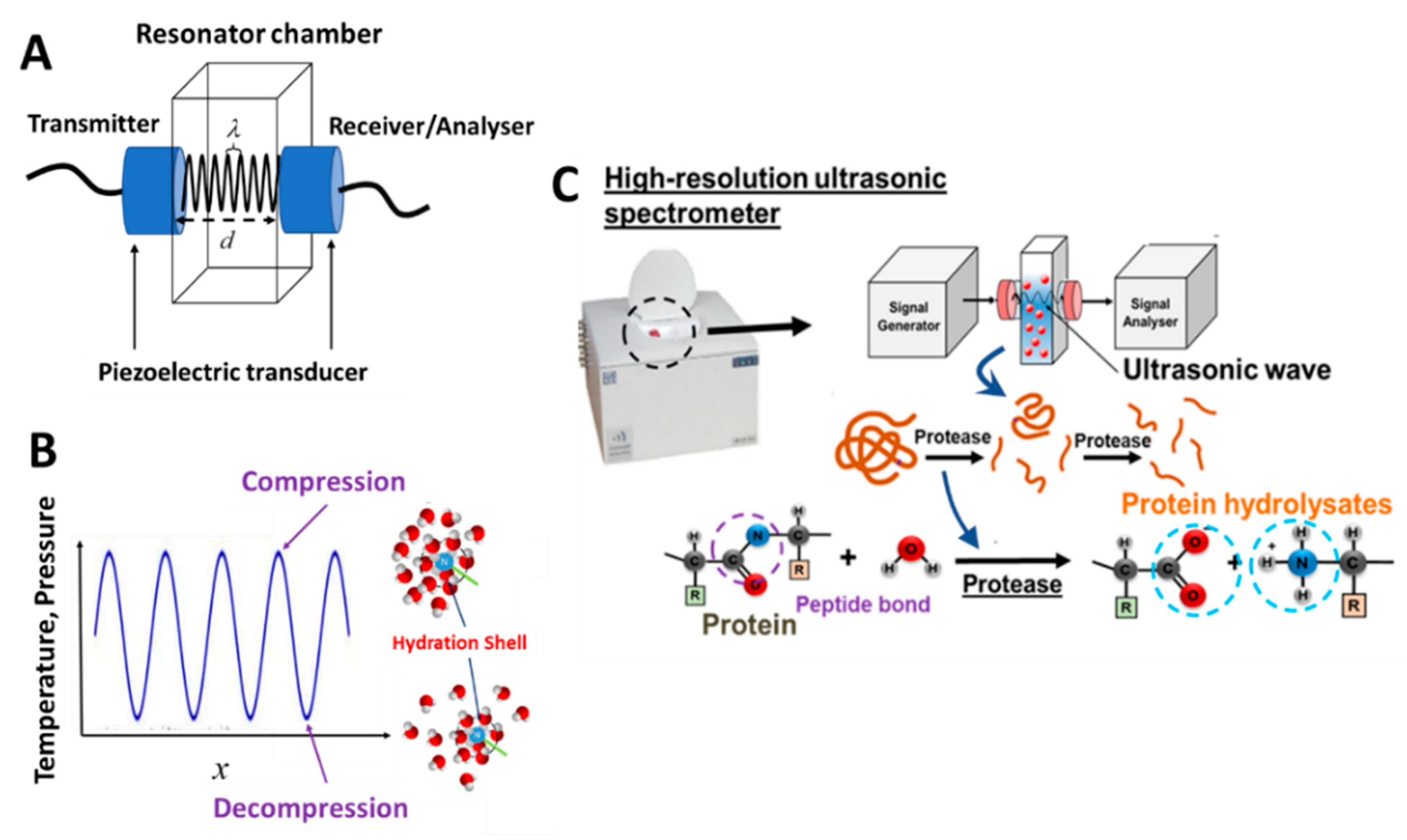
4. High-Resolution Ultrasonic Spectroscopy (HR-US)
4.1. Principles of HR-US
4.2. Key Advancement and Attributes of HR-US
4.3. Kinetics of Protein Hydrolysis
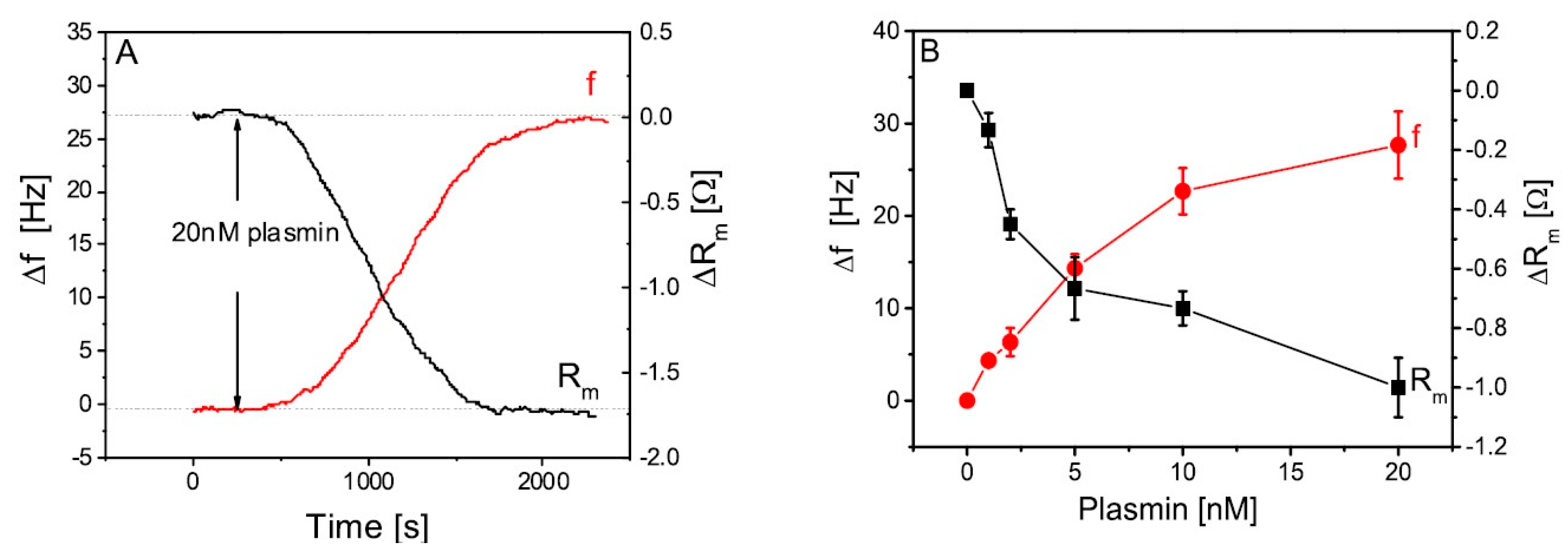
4.4. Detection of Proteases Activity by HR-US
4.5. Examples of Ultrasonic Measurements of Proteolysis
| Protease | Reactions and Conditions | Reference |
|---|---|---|
| Chymosin | Ultrasonic renneting process of milk at 30 °C. Ultrasonic analysis of multistage structural rearrangement. | [113] |
| Chymosin | Ultrasonic renneting process of milk at 30 °C. Comparison of effect of pH, temperature and enzyme-induced gelation in milks. | [114] |
| Chymosin | Ultrasonic renneting process of milk at 30 °C. Effect of preheat treatment at ultra-high temperature. | [116] |
| Proteinase K | Hydrolysis of Gly-Leu-Gly-Gly-Ala (synthetic pentapeptides) in 30-mM Tris buffer, pH 8, at 37 °C. Effect of substrate-enzyme ratio. | [109,119] |
| Proteinase K | Hydrolysis of bovine serum albumin (BSA) in 30-mM Tris buffer, pH 8, at 37 °C. Effect of enzyme concentration. | [109,119] |
| Proteinase K | Hydrolysis of bovine casein aggregates in 30-mM Tris buffer, pH 8, at 37 °C. Effect of substate concentration. | [109,119] |
| Chymosin (Maxiren® 180) | Proteolytic activity in milk samples at 30 °C. Screening method of enzyme activity. | [117] |
| Protease (Bakezyme B500®) a | Proteolytic activity in wheat flour suspension at 30 °C. Screening method of enzyme activity. | [117] |
| Trypsin | Hydrolysis of β-casein in 50-mM phosphate, pH 7.5, at 37 °C | [111] |
| Pancreatin | Proteolysis of casein solution at 37 °C. Effect of hydrolytic conditions relevant to pharmacopoeia assays. | [118] |
| α-chymotrypsin | Hydrolysis of β-lactoglobulin in 0.1-mol kg−1 phosphate buffer, pH 7.8, at 25 °C. Effect of substrate concentration. | [73] |
| Trypsin | Hydrolysis of β-casein in 0.1-mol kg−1 phosphate buffer. Effect of pH (6.6–8), temperature (15–45 °C) and trypsin concentration. | [104] |
4.6. Future Works and Limitations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Grufferty, M.B.; Fox, P.F. Milk alkaline proteinase. J. Dairy Res. 1988, 55, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Bastian, E.D.; Brown, R.J. Plasmin in milk and dairy products: An update. Int. Dairy J. 1996, 6, 435–457. [Google Scholar] [CrossRef]
- Abdul, S.; Leebeek, F.W.G.; Rijken, D.C.; de Willige, S.U. Natural heterogeneity of α2-antiplasmin: Functional and clinical consequences. Blood 2016, 127, 538–545. [Google Scholar] [CrossRef] [Green Version]
- Dupont, D.; Bailly, C.; Grosclaude, J.; Collin, J.C. Differential titration of plasmin and plasminogen in milk using sandwich ELISA with monoclonal antibodies. J. Dairy Res. 1997, 64, 77–86. [Google Scholar] [CrossRef]
- Rauh, V.M.; Johansen, L.B.; Ipsen, R.; Paulsson, M.; Larsen, L.B.; Hammershøj, M. Plasmin activity in UHT milk: Relationship between proteolysis, age gelation, and bitterness. J. Agricult. Food Chem. 2014, 62, 6852–6860. [Google Scholar] [CrossRef]
- Arena, S.; Salzano, A.M.; Scalon, A. Identification of protein markers for the occurrence of defrosted material in milk through a MALDI-TOF-MS profiling approach. J. Proteom. 2016, 147, 56–65. [Google Scholar] [CrossRef]
- Volitaki, A.; Kaminarides, S. Detection of bovine milk in ovine Halloumi cheese by HPLC analysis of cheese caseins hydrolysed by plasmin. Milchwissenschaft 2001, 56, 207–210. [Google Scholar]
- Miralles, B.; Ramos, M.; Amigo, L. Influence of proteolysis of milk on the whey protein to total protein ratio as determined by capillary electrophoresis. J. Dairy Sci. 2003, 86, 2813–2817. [Google Scholar] [CrossRef] [Green Version]
- Walstra, P.; Wouters, J.T.M.; Geurts, T.J. Dairy Science and Technology, 2nd ed.; CRC Press: Boca Raton, FL, USA; Taylor and Francis Group: Oxford, UK, 2005; pp. 17–108. [Google Scholar]
- Raynal-Ljutovac, K.; Pirisi, A.; Cremoux, R.; Gonzalo, C. Somatic cells of goat and sheep milk: Analytical, sanitary, productive and technological aspects. Small Rumin. Res. 2007, 68, 126–144. [Google Scholar] [CrossRef]
- Schwarz, D.; Diesterbeck, U.S.; Konig, S.; Brugemann, K.; Schlez, K.; Zschock, M.; Wolter, W.; Czerny, C.P. Flow cytometric differential cell counts in milk for the evaluation of inflammatory reactions in clinically healthy and subclinically infected bovine mammary glands. J. Dairy Sci. 2011, 94, 5033–5044. [Google Scholar] [CrossRef] [Green Version]
- Muller-Renaud, S.; Dupont, D.; Dulieu, P. Quantification of β-casein in milk and cheese using an optical immunosensor. J. Agric. Food Chem. 2004, 52, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Dallas, D.C.; Murray, N.M.; Gan, J. Proteolytic systems in milk: Perspectives on the evolutionary function within the mammary gland and the infant. J. Mammary Gland Biol. Neoplasia 2015, 20, 133–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halabi, A.; Deglaire, A.; Hennetier, M.; Violleau, F.; Burel, A.; Bouhallab, S.; Dupond, D.; Croguennec, T. Structural characterization of heat-induced protein aggregates in model infant milk formulas. Food Hydrocol. 2020, 107, 105928. [Google Scholar] [CrossRef]
- Bumberger, E.; Belitz, H.D. Bitter taste of enzymic hydrolysates of casein. I. Isolation, structural and sensorial analysis of peptides from tryptic hydrolysates of β-casein. Z. Lebensm. Unters Forsch. 1993, 197, 14–19. [Google Scholar] [CrossRef]
- Drinane, M.; Sherman, J.A.; Hall, A.E.; Simons, M.; Mulligan-Kehoe, M.J. Plasminogen and plasmin activity in patients with coronary artery disease. J. Thromb. Haemost. 2006, 4, 1288–1295. [Google Scholar] [CrossRef]
- Cesarman-Maus, G.; Hajjar, K.A. Molecular mechanisms of fibrinolysis. Br. J. Haematol. 2005, 129, 307–321. [Google Scholar] [CrossRef]
- Heegaard, C.W.; Amdreasen, P.; Petersen, T.R.; Ramussen, L.K.I. Binding of plasminogen and tissue-type plasminogen activator to dimeric αs2-casein accelerates plasmin generation. Fibrinolysis Proteolysis 1997, 11, 29–36. [Google Scholar] [CrossRef]
- Politis, I.; White, J.H.; Zavizion, B.; Goldberg, J.J.; Guo, M.R.; Kindstedt, P. Effect of individual caseins on plasminogen activation by bovine urokinase-type and tissue-type plasminogen activators. J. Dairy Sci. 1995, 78, 484–490. [Google Scholar] [CrossRef]
- Rippel, K.M.; Nielson, S.S.; Hayes, K.D. Effects of native and denatured whey proteins on plasminogen activator activity. J. Dairy Sci. 2004, 87, 2344–2350. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.L. Indigenous proteinases in milk. In Advanced Dairy Chemistry Volume 1, Proteins; Fox, P.F., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2003; pp. 495–521. [Google Scholar]
- Deeth, H.C.; Datta, N. Heat Treatment of Milk|Ultra-High Temperature Treatment (UHT): Heating Systems. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., McSweeney, P.L.H., Fox, P.F., Eds.; Elsiever Science Publishing Co Inc.: San Diego, CA, USA, 2011; pp. 732–737. [Google Scholar]
- Newstead, D.F.; Paterson, G.; Anema, S.G.; Coker, C.J.; Wewala, A.R. Plasmin activity in direct-steam-injection UHT-processed reconstituted milk: Effects of preheat treatment. Int. Dairy J. 2006, 16, 573–579. [Google Scholar] [CrossRef]
- Saint Denis, T.; Humbert, G.; Gaillard, J. Heat inactivation of native plasmin, plasminogen and plasminogen activators in bovine milk: A revisited study. Le Lait 2001, 81, 715–729. [Google Scholar] [CrossRef]
- Lu, D.D.; Nielsen, S.S. Heat inactivation of native plasminogen activators in bovine milk. J. Food Sci. 1993, 58, 1010–1016. [Google Scholar] [CrossRef]
- Kennedy, A.; Kelly, A.L. The influence of somatic cell count on the heat stability of bovine milk plasmin activity. Int. Dairy J. 1997, 7, 717–721. [Google Scholar] [CrossRef]
- Hayes, M.G.; McSweeney, P.L.H.; Kelly, A.L. The influence of native and heat denatured whey proteins on enzyme activity. 1. Plasmin. Milchwissenschaft 2002, 57, 208–211. [Google Scholar]
- Monti, J.C.; Mermoud, A.F.; Jollès, P. Trypsin in human milk. Experientia 1986, 42, 39–41. [Google Scholar] [CrossRef]
- McGilligan, K.M.; Thomas, D.W.; Eckhert, C.D. Alpha-1-antitrypsin concentration in human milk. Pediatric Res. 1987, 22, 268–270. [Google Scholar] [CrossRef] [Green Version]
- Magboul, A.A.; Larsen, L.B.; McSweeney, P.L.; Kelly, A.L. Cysteine protease activity in bovine milk. Int. Dairy J. 2001, 11, 865–872. [Google Scholar] [CrossRef]
- Palmer, D.J.; Kelly, V.C.; Smit, A.M.; Kuy, S.; Knight, C.G.; Cooper, G.J. Human colostrum: Identification of minor proteins in the aqueous phase by proteomics. Proteomics 2006, 6, 2208–2216. [Google Scholar] [CrossRef]
- Fiorillo, A.S.; Critello, C.D.; Pullano, S.A. Theory, technology and applications of piezoresistive sensors: A review. Sens. Actuators A 2018, 281, 156–175. [Google Scholar] [CrossRef]
- Fiorillo, A.S.; Pullano, S.A.; Bianco, M.G.; Critello, C.D. Ultrasonic Transducers Shaped in Archimedean and Fibonacci Spiral: A Comparison. Sensors 2020, 20, 2800. [Google Scholar] [CrossRef]
- Bundle, R.L.; Jarvi, E.J.; Rosentreter, J.J. Piezoelectric quartz crystal biosensors. Talanta 1998, 46, 1223–1236. [Google Scholar] [CrossRef]
- Clark, L.C.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Thévenit, D.R.; Toth, K.; Durst, R.A.; Wolson, G.S. Electrochemical biosensors: Recommended definitions and classifications. Biosens. Bioelectr. 2001, 16, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.Q.; Luo, J.K.; Nguyen, N.T.; Walton, A.J.; Flewitt, A.J.; Zu, X.T.; Li, Y.; McHale, G.; Matthews, A.; Iborra, E.; et al. Advances in piezoelectric thin films for acoustic biosensors, acousto fluidics and lab-on-chip applications. Prog. Mat. Sci. 2017, 89, 31–91. [Google Scholar] [CrossRef] [Green Version]
- Manbachi, A.; Cobbold, R.S.C. Development and application of piezoelectric materials for ultrasound generation and detection. Ultrasound 2011, 19, 187–196. [Google Scholar] [CrossRef]
- Howells, C.H. Piezoelectric energy harvesting. Energy Convers. Manag. 2009, 50, 1847–1850. [Google Scholar] [CrossRef]
- Sauerbrey, G. Verwendung von schwingquarzen zur wagung dunnerschichten und zur mikrowagung. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Zainuddin, A.A.; Nordin, A.N.; Rahim, R.A.; Ralib, A.A.M.; Khan, S.; Guines, C.; Chatras, M.; Pothier, A. Verification of quartz crystal microbalance array using vector network analyzer and open QCM. Indonesian J. Electrical Eng. Comput. Sci. 2018, 10, 84–93. [Google Scholar] [CrossRef]
- Buttry, A.; Ward, M.D. Measurement of interfacial processes at electrode surfaces with the electrochemical quartz crystal microbalance. Chem. Rev. 1992, 6, 1355–1379. [Google Scholar] [CrossRef]
- Lu, C. Theory and practice of the quartz crystal microbalance. In Applications of Piezoelectric Quartz Crystal Microbalances, 2nd ed.; Lu, C., Czanderna, A.W., Eds.; Elsiever Science Publishers B.V.: Amsterdam, The Netherlands, 1984; Volume 7, pp. 19–62. [Google Scholar]
- Ballantine, D.S.; White, R.M.; Martin, S.J.; Ricco, A.J.; Zellers, E.T.; Frye, G.C.; Wohltjen, H. Acoustic Wave Sensors. Theory, Design, and Physico-Chemical Applications; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Rodahl, M.; Kasemo, F.H. QCM operation in liquids: An explanation of measured variations in frequency and Q factor with liquid conductivity. Anal. Chem. 1996, 68, 2219–2227. [Google Scholar] [CrossRef]
- Jonsson, M.P.; Jönsson, P.; Höök, F. Simultaneous nanoplasmonic and quartz crystal microbalance sensing: Analysis of biomolecular conformational changes and quantification of the bound molecular mass. Anal. Chem. 2008, 80, 7988–7995. [Google Scholar] [CrossRef] [PubMed]
- McNamara, T.P.; Blandford, C.F. A sensitivity metric and software to guide the analysis of soft films measured by a quartz crystal microbalance. Analyst 2016, 141, 2911–2919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohanka, M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitakis, M.; Gizeli, E. Acoustic sensors as a biophysical tool for probing cell attachment and cell/surface interactions. Cell. Molec. Life Sci. 2012, 69, 357–371. [Google Scholar] [CrossRef]
- Elmlund, L.; Käck, C.; Aastrup, T.; Nicholls, I.A. Study of the interaction of trastuzumab and SKOV3 epithelial cancer cells using a quartz crystal microbalance sensor. Sensors 2015, 15, 5884–5894. [Google Scholar] [CrossRef] [Green Version]
- Afzal, A.; Mujahid, A.; Schirhagl, R.; Bajwa, S.Z.; Latif, U.; Feroz, S. Gravimetric viral diagnostics: QCM based biosensors for early detection of viruses. Chemosensors 2017, 5, 7. [Google Scholar] [CrossRef]
- Thompson, M.; Ballantyne, S.M.; Cheran, L.E.; Stevenson, A.C.; Loweb, C.H.R. Electromagnetic excitation of high frequency acoustic waves and detection in the liquid phase. Analyst 2003, 128, 1048–1055. [Google Scholar] [CrossRef]
- Sheikh, S.; Blaszykowski, C.; Thompson, M. Sacrificial BSA to block non-specific adsorption on organosilane adlayers in ultra-high frequency acoustic wave sensing. Surf. Interface Anal. 2013, 45, 1781–1784. [Google Scholar] [CrossRef]
- Neves, M.A.D.; Blaszykowski, C.; Bokhari, S.; Thompson, M. Ultrahigh frequency piezoelectric aptasensor for the label-free detection of cocaine. Biosens. Bioelectr. 2015, 72, 383–392. [Google Scholar] [CrossRef]
- Sheikh, S.; Blaszykowski, C.; Thompson, M. Label-free detection of HIV-2 antibodies in serum with an ultra-high frequency acoustic wave sensor. Talanta 2011, 85, 816–819. [Google Scholar] [CrossRef]
- Crivianu-Gaita, V.; Aamer, M.; Posaratnanathan, R.T.; Romaschin, A.; Thompson, M. Acoustic wave biosensor for the detection of the breast and prostate cancer metastasis biomarker protein PTHrP. Biosens. Bioelectr. 2016, 78, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, S.; De La Franier, B.; Hianik, T.; Thompson, M. Surface probe linker with tandem anti-fouling properties for application in biosensor technology. Biosensors 2020, 10, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strube, O.I.; Rüdiger, A.A.; Bremser, W. Buildup of biobased adhesive layers by enzymatically controlled deposition on the example of casein. Int. J. Ahesion Ahesives 2015, 63, 9–13. [Google Scholar] [CrossRef]
- Matsunage, Y.; Suenaga, M.; Takahashi, H.; Furuta, A. Vitamin D affects neuronal peptides in neurodegenerative disease: Differences of V-D2 and V-D3 for affinity to amyloid-β and scrapie prion protein in vitro. In A Critical Evaluation of Vitamin D—Clinical Overview; Sivakumar, J.T.G., Ed.; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar]
- Poturnayova, A.; Castillo, G.; Subjakova, V.; Tatarko, M.; Snejdarkova, M.; Hianik, T. Optimization of the cytochrome c detection by acoustic and electrochemical methods based on aptamer sensors. Sens. Actuators B 2017, 228, 817–827. [Google Scholar] [CrossRef]
- Chen, J.B.; Neves, M.A.D.; Thompson, M. Biosensor surface attachment of the ovarian cancer biomarker HSP10 via His-tag modification. Sens. Bio Sens. Res. 2016, 11, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Tatarko, M.; Muckley, E.; Šubjaková, V.; Goswami, M.; Sumpter, B.; Hianik, T.; Ivanov, I.N. Machine learning enabled acoustic detection of subnanomolar concentration of trypsin and plasmin in solution. Sens. Actuators B 2018, 272, 282–288. [Google Scholar] [CrossRef]
- Poturnayova, A.; Karpisova, I.; Castillo, G.; Mező, G.; Kocsis, L.; Csámpai, A.; Keresztes, Z.; Hianik, T. Detection of plasmin based on specific peptide substrate usingacoustic transducer. Sens. Actuators B 2016, 223, 591–598. [Google Scholar] [CrossRef]
- Románski, L.; Tatarko, M.; Jiao, M.; Keresztes, Z.; Hianik, T.; Thompson, M. Casein probe-based fast plasmin determination in the picomolar range by an ultra-high frequency acoustic wave biosensor. Sens. Actuators B 2018, 275, 206–214. [Google Scholar] [CrossRef]
- Sabot, A.; Krause, S. Simultaneous quartz crystal microbalance impedance and electrochemical impedance measurements. Investigation into the degradation of thin polymer films. Anal. Chem. 2002, 74, 3304–3311. [Google Scholar] [CrossRef]
- Stair, L.J.; Watkinson, M.; Krause, S. Sensor materials for detection of proteases. Biosens. Bioelectr. 2009, 24, 2113–2118. [Google Scholar] [CrossRef]
- Ahmad, N.; Colak, B.; Zhang, D.-W.; Gibbs, M.J.; Watkinson, M.; Becer, C.R.; Gautrot, J.E.; Krause, S. Peptide cross-linked poly (ethylene glycol) hydrogel films as biosensor coatings for the detection of collagenase. Sensors 2019, 19, 1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huenerbein, A.; Schmelzer, C.E.H.; Neubert, R.H.H. Real-time monitoring of peptic and tryptic digestions of bovine β-casein using quartz crystal microbalance. Anal. Chim. Acta 2007, 584, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.; Ho, Y.H.; Sung, T.C.; Wu, W.F.; Chen, C.S. Escherichia coli proteome microarrays identified the substrates of ClpYQ protease. Mol. Cell. Proteom. 2017, 16, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Ong, I.L.H.; Yang, K.L. Recent developments in protease activity assays and sensors. Analyst 2017, 142, 1867–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckin, V. High-resolution ultrasonic spectroscopy. J. Sens. Sens. Syst. 2018, 7, 207–217. [Google Scholar] [CrossRef]
- Elvira, L.; Resa, P.; Castro, P.; Shukla, S.K.; Sierra, C.; Aparicio, C.; Durán, C.; de Espinosa, F.M. Bioprocess Monitoring using Low-intensity Ultrasound. In Ultrasound in Food Processing: Recent Advances; Villamiel, M., Montilla, A., García-Pérez, J.V., Cárcel, J.A., Benedito, J., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2017; pp. 146–174. [Google Scholar]
- Buckin, V.; Altas, M.C. Ultrasonic monitoring of biocatalysis in solutions and complex dispersions. Catalysts 2017, 7, 336. [Google Scholar] [CrossRef] [Green Version]
- Kaatze, U.; Eggers, F.; Lautscham, K. Ultrasonic velocity measurements in liquids with high resolution—Techniques, selected applications and perspectives. Meas. Sci. Technol. 2008, 19, 062001. [Google Scholar] [CrossRef]
- Kudryashov, E.; Smyth, C.; Duffy, G.; Buckin, V. Ultrasonic high-resolution longitudinal and shear wave measurements in food colloids: Monitoring of gelation processes and detection of pathogens. Trends Colloid Interface Sci. Xiv 2000, 115, 287–294. [Google Scholar]
- Povey, M.J.W. Ultrasonic Techniques for Fluids Characterization, 1st ed.; Academic Press: San Diego, CA, USA, 1997; pp. 1–214. [Google Scholar]
- Herzfeld, K.F.; Litovitz, T.A. Absorption and Dispersion of Ultrasonic Waves, 1st ed.; Academic Press: New York, NY, USA, 1959; pp. 1–536. [Google Scholar]
- Buckin, V. Application of high-resolution ultrasonic spectroscopy for analysis of complex formulations. Compressibility of solutes and solute particles in liquid mixtures. IOP Conf. Ser. Mater. Sci. Eng. 2012, 42, 012001. [Google Scholar] [CrossRef]
- Smyth, C.; Kudryashov, E.D.; Buckin, V. High-frequency shear and volume viscoelastic moduli of casein particle gel. Colloids Surf. A 2001, 183, 517–526. [Google Scholar] [CrossRef]
- Audebrand, M.; Doublier, J.L.; Durand, D.; Emery, J.R. Investigation of gelation phenomena of some polysaccharides by ultrasonic spectroscopy. Food Hydrocoll. 1995, 9, 195–203. [Google Scholar] [CrossRef]
- Buckin, V.; O’ Driscoll, B. Ultrasonic waves and material analysis: Recent advances and future trends. LabPlus Int. 2002, 16, 17–21. [Google Scholar]
- Buckin, V.; O’ Driscoll, B.; Smyth, C. Ultrasonic spectroscopy for material analysis. Recent advances. Spectrosc. Eur. 2003, 15, 20–25. [Google Scholar]
- Buckin, V.; Hallone, S. Ultrasonic characterisation of w/o microemulsions—Structure, phase diagrams, state of water in nano-droplets, encapsulated proteins, enzymes. In Microemulsions—An Introduction to Properties and Applications; Najjar, R., Ed.; InTech: Rijeka, Croatia, 2012; pp. 33–66. [Google Scholar]
- Del Grosso, V.A.; Mader, C.W. Speed of sound in pure water. J. Acoust. Soc. Am. 1972, 52, 1442–1446. [Google Scholar] [CrossRef]
- Wubshet, S.G.; Måge, I.; Böcker, U.; Lindberg, D.; Knutsen, S.H.; Rieder, A.; Rodriguez, D.A.; Afseth, N.K. FTIR as a rapid tool for monitoring molecular weight distribution during enzymatic protein hydrolysis of food processing by-products. Anal. Methods 2017, 9, 4247–4254. [Google Scholar] [CrossRef]
- Vorob’ev, M.M.; Kochetkov, K.A. Determination of kinetic parameters for casein hydrolysis by chymotrypsin using two ranges of substrate concentration. Int. Dairy J. 2016, 61, 76–84. [Google Scholar] [CrossRef]
- Valencia, P.; Pinto, M.; Almonacid, S. Identification of the key mechanisms involved in the hydrolysis of fish protein by Alcalase. Process Biochem. 2014, 49, 258–264. [Google Scholar] [CrossRef]
- Márquez, M.C.; Vázquez, M.A. Modeling of enzymatic protein hydrolysis. Process Biochem. 1999, 35, 111–117. [Google Scholar] [CrossRef]
- Tardioli, P.W.; Sousa, R.; Giordano, R.C.; Giordano, R.L.C. Kinetic model of the hydrolysis of polypeptides catalyzed by Alcalase® immobilized on 10% glyoxyl-agarose. Enzym. Microb. Technol. 2005, 36, 555–564. [Google Scholar] [CrossRef]
- Vorob’ev, M.M. Kinetics of peptide bond demasking in enzymatic hydrolysis of casein substrates. J. Mol. Catal. B Enzym. 2009, 58, 146–152. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Enzymic Hydrolysis of Food Protein; Elsevier Applied Science: London, UK, 1986; Volume 8, pp. 1–427. [Google Scholar]
- Rutherfurd, S.M. Methodology for determining degree of hydrolysis of proteins in hydrolysates: A Review. J. AOAC Int. 2010, 93, 1515–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellman, D.; McEvoy, E.; O’Cuinn, G.; FitzGerald, R. Proteinase and exopeptidase hydrolysis of whey protein: Comparison of the TNBS, OPA and pH stat methods for quantification of degree of hydrolysis. Int. Dairy J. 2003, 13, 447–453. [Google Scholar] [CrossRef]
- Beaubier, S.; Framboisier, X.; Ioannou, I.; Galet, O.; Kapel, R. Simultaneous quantification of the degree of hydrolysis, protein conversion rate and mean molar weight of peptides released in the course of enzymatic proteolysis. J. Chromatogr. B 2019, 1105, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Camacho, F.; González-Tello, P.; Páez-Dueñas, M.-P.; Guadix, E.-M.; Guadix, A. Correlation of base consumption with the degree of hydrolysis in enzymic protein hydrolysis. J. Dairy Res. 2001, 68, 251–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodin, A.; Framboisier, X.; Alonso, D.; Marc, I.; Kapel, R. Size-exclusion HPLC as a sensitive and calibrationless method for complex peptide mixtures quantification. J. Chromatogr. B 2015, 1006, 71–79. [Google Scholar] [CrossRef]
- Kristoffersen, K.A.; Afseth, N.K.; Böcker, U.; Lindberg, D.; de Vogel-van den Bosch, H.; Ruud, M.L.; Wubshet, S.G. Average molecular weight, degree of hydrolysis and dry-film FTIR fingerprint of milk protein hydrolysates: Intercorrelation and application in process monitoring. Food Chem. 2020, 310, 125800. [Google Scholar] [CrossRef]
- Kristoffersen, K.A.; Liland, K.H.; Böcker, U.; Wubshet, S.G.; Lindberg, D.; Horn, S.J.; Afseth, N.K. FTIR-based hierarchical modeling for prediction of average molecular weights of protein hydrolysates. Talanta 2019, 205, 120084. [Google Scholar] [CrossRef]
- Lynch, R.; Burke, A.; Byrne, J.; Buckin, V. Osmolality and molar mass of oligosaccharides in breast milks and infant formula during hydrolysis of lactose. Application of high-resolution ultrasonic spectroscopy. Food Chem. 2020, 322, 126645. [Google Scholar] [CrossRef]
- Luo, Q.; Chen, D.; Boom, R.M.; Janssen, A.E.M. Revisiting the enzymatic kinetics of pepsin using isothermal titration calorimetry. Food Chem. 2018, 268, 94–100. [Google Scholar] [CrossRef]
- Cheison, S.C.; Kulozik, U. Impact of the environmental conditions and substrate pre-treatment on whey protein hydrolysis: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 418–453. [Google Scholar] [CrossRef]
- Gonzàlez-Tello, P.; Camacho, F.; Jurado, E.; Páez, M.P.; Guadix, E.M. Enzymatic hydrolysis of whey proteins: I. Kinetic models. Biotechnol. Bioeng. 1994, 44, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Marquez Moreno, M.C.; Fernandez Cuadrado, V. Enzymic hydrolysis of vegetable proteins: Mechanism and kinetics. Process Biochem. 1993, 28, 481–490. [Google Scholar] [CrossRef]
- Melikishvili, S.; Dizon, M.; Hianik, T. Application of high-resolution ultrasonic spectroscopy for real-time monitoring of trypsin activity in β-casein solution. Food Chem. 2021, 337, 127759. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.M.; Danson, M.J. A new understanding of how temperature affects the catalytic activity of enzymes. Trends Biochem. Sci. 2010, 35, 584–591. [Google Scholar] [CrossRef]
- Arcus, V.L.; Prentice, E.J.; Hobbs, J.K.; Mulholland, A.J.; Van der Kamp, M.W.; Pudney, C.R.; Parker, E.J.; Schipper, L.A. On the temperature dependence of enzyme-catalyzed rates. Biochemistry 2016, 55, 1681–1688. [Google Scholar] [CrossRef]
- Resa, P.; Buckin, V. Ultrasonic analysis of kinetic mechanism of hydrolysis of cellobiose by beta-glucosidase. Anal. Biochem. 2011, 415, 1–11. [Google Scholar] [CrossRef]
- Altas, M.C.; Kudryashov, E.; Buckin, V. Ultrasonic monitoring of enzyme catalysis; enzyme activity in formulations for lactose-intolerant infants. Anal. Chem. 2016, 88, 4714–4723. [Google Scholar] [CrossRef]
- Vorob’ev, M.M.; Vogel, V.; Güler, G.; Mäntele, W. Monitoring of demasking of peptide bonds during proteolysis by analysis of the apparent spectral shift of intrinsic protein fluorescence. Food Biophys. 2011, 6, 519–526. [Google Scholar] [CrossRef]
- Buckin, V.; Craig, E. New applications of high-resolution ultrasonic spectroscopy for real-time analysis of enzymatic proteolysis. Eur. Biopharm. Rev. (Summer) 2005, 50–53. [Google Scholar]
- Born, K.; Manns, A.; Dzeyk, K.; Lutz-Wahl, S.; Gau, D.; Fischer, L. Evaluation of ultrasound velocity measurements for estimating protease activities using casein as substrate. Biotechnol. Lett. 2010, 32, 249–253. [Google Scholar] [CrossRef]
- Taulier, N.; Chalikian, T.V. Characterization of pH-induced transitions of beta-lactoglobulin: Ultrasonic, densimetric, and spectroscopic studies. J. Mol. Biol. 2001, 314, 873–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwyer, C.; Donnelly, L.; Buckin, V. Ultrasonic analysis of rennet-induced pre-gelation and gelation processes in milk. J. Dairy Res. 2005, 72, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Corredig, M.; Alexander, M.; Dalgleish, D.G. The application of ultrasonic spectroscopy to the study of the gelation of milk components. Food Res. Int. 2004, 37, 557–565. [Google Scholar] [CrossRef]
- Bakkali, F.; Moudden, A.; Faiz, B.; Amghar, A.; Maze, G.; Espinosa, F.M.; Akhnak, M. Ultrasonic measurement of milk coagulation time. Meas. Sci. Technol. 2001, 12, 2154–2159. [Google Scholar] [CrossRef]
- Wang, Q.; Bulca, S.; Kulozik, U. A comparison of low-intensity ultrasound and oscillating rheology to assess the renneting properties of casein solutions after UHT heat pre-treatment. Int. Dairy J. 2007, 17, 50–58. [Google Scholar] [CrossRef]
- Dijk, A.A.V.; Tilborg, M.W.E.M.V.; Gerritsma, J.S.J.; Olsthoorn, M.M.A.; Folkertsma, B.; Westerlaken, I. Novel Method for Screening and Selecting Enzymes. 2007. Patent. Available online: http://europepmc.org/article/PAT/EP1761636 (accessed on 17 June 2020).
- Niemeyer, K. Ultrasonic Methods for Analytical Determination of Pancreatic Enzyme Activities in Pharmaceutical Preparations. Ph.D. Thesis, Technische Universität Braunschweig, Braunschweig, Germany, 2012. [Google Scholar]
- Craig, E. Detection and Partial Characterization of Biopolymer Aggregates Using High Resolution Ultrasonic Spectroscopy. Ph.D. Thesis, National University of Ireland, University College Dublin, Dublin, Ireland, 2004. [Google Scholar]
- Dizon, M.; Tatarko, M.; Szabo, K.; Hianik, T. Comparative analysis of plasmin detection by high resolution ultrasonic spectroscopy and ELISA in milk samples. Food Chem. 2020. under review. [Google Scholar]
- Poturnayova, A.; Szabo, K.; Hucker, A.; Kocsis, R.; Hianik, T. Determination of plasmin in milk using QCM and ELISA methods. Food Control 2020. under review. [Google Scholar]





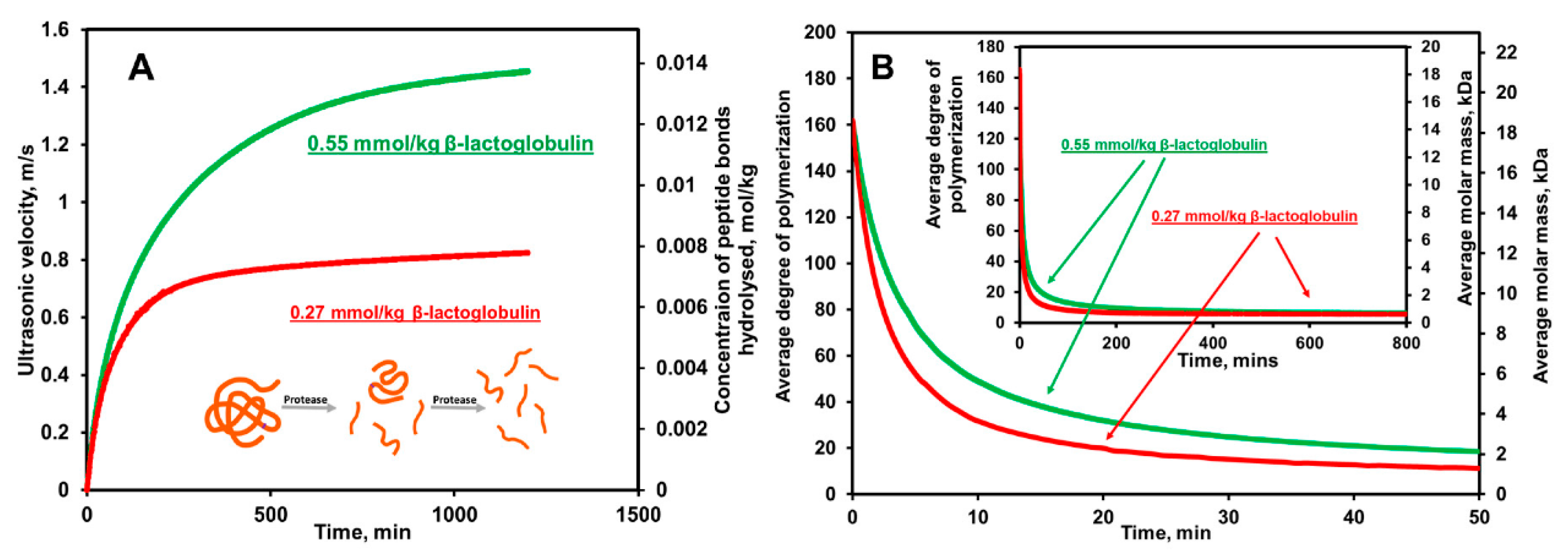
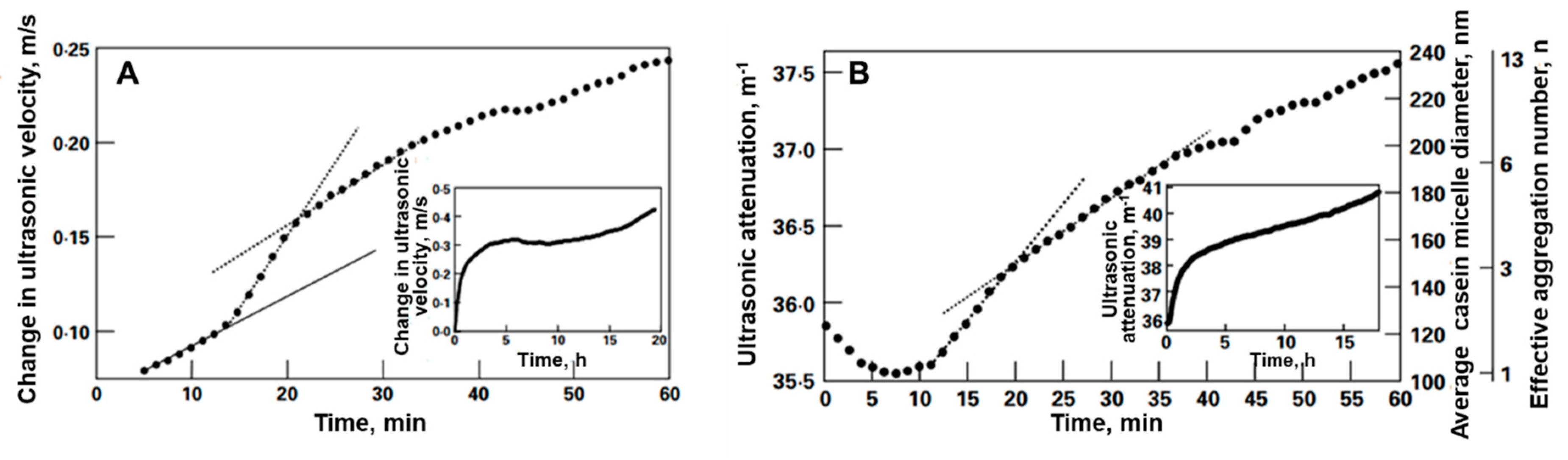
| Acoustic Method | Detection Phase | Substrate | Limit of Detection | Time of Detection | Reference |
|---|---|---|---|---|---|
| QCM | surface | Short peptides at gold surface | 0.65 nM Plasmin | 30 min | [63] |
| QCM | surface | β-casein at gold substrate | 0.1 nM Trypsin 1 nM Plasmin | 30 min | [62] |
| EMPAS | surface | β-casein at gold substrate | 0.032 nM Plasmin | 30 min | [64] |
| HR-US | volume | 0.1–1% (w/w) β-casein in buffer | 0.2 nM Trypsin | 15–30 min | [104] |
| HR-US | volume | 0.1–1% (w/w) β-casein in buffer | 0.2 nM Plasmin | 15–30 min | [120] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dizon, M.; Tatarko, M.; Hianik, T. Advances in Analysis of Milk Proteases Activity at Surfaces and in a Volume by Acoustic Methods. Sensors 2020, 20, 5594. https://doi.org/10.3390/s20195594
Dizon M, Tatarko M, Hianik T. Advances in Analysis of Milk Proteases Activity at Surfaces and in a Volume by Acoustic Methods. Sensors. 2020; 20(19):5594. https://doi.org/10.3390/s20195594
Chicago/Turabian StyleDizon, Mark, Marek Tatarko, and Tibor Hianik. 2020. "Advances in Analysis of Milk Proteases Activity at Surfaces and in a Volume by Acoustic Methods" Sensors 20, no. 19: 5594. https://doi.org/10.3390/s20195594






