Reduce Calibration Time in Motor Imagery Using Spatially Regularized Symmetric Positives-Definite Matrices Based Classification
Abstract
1. Introduction
2. Geometry of SPD Matrices
- i.e., SPD matrices are invertible.
- , eigenvalues are positive i.e., .
2.1. Riemannian Natural Manifold
2.2. Riemannian Distance
2.3. Riemannian Mean (Choice of Reference Point)
2.4. Minimum Distance to Riemannian Mean (MDRM)
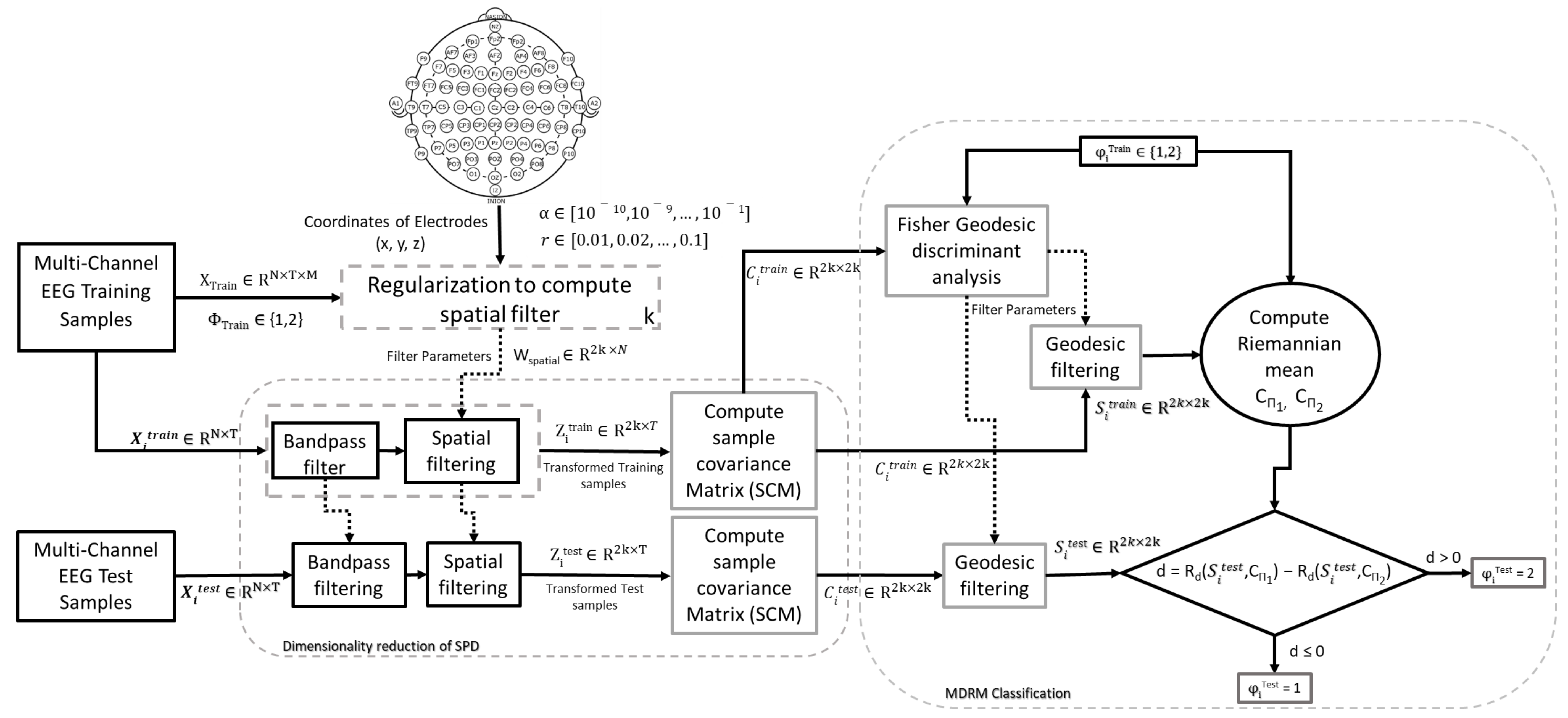
3. Methodology
4. Data and Experiment
4.1. Dataset IVa, BCI Competition III
4.2. Dataset IIIa, BCI Competition III
4.3. Dataset IIa, BCI Competition IV
4.4. Experimental Setup
4.5. Evaluation Metrics
5. Results and Discussion
5.1. Dataset IVa, BCI Competition III
5.2. Dataset IIIa, BCI Competition III
5.3. Dataset IIa, BCI Competition IV
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, A.; Lal, S.; Guesgen, H.W. Architectural Review of Co-Adaptive Brain Computer Interface. In Proceedings of the 2017 4th Asia-Pacific World Congress on Computer Science and Engineering, Nadi, Fiji, 11–13 December 2017; pp. 200–207. [Google Scholar]
- Blankertz, B.; Dornhege, G.; Krauledat, M.; Müller, K.R.; Curio, G. The non-invasive Berlin Brain–Computer Interface: Fast acquisition of effective performance in untrained subjects. NeuroImage 2007, 37, 539–550. [Google Scholar] [CrossRef]
- Ramadan, R.A.; Vasilakos, A.V. Brain computer interface: Control signals review. Neurocomputing 2017, 223, 26–44. [Google Scholar] [CrossRef]
- Barachant, A.; Bonnet, S.; Congedo, M.; Jutten, C. Classification of covariance matrices using a Riemannian-based kernel for BCI applications. Neurocomputing 2013, 112, 172–178. [Google Scholar] [CrossRef]
- Thomas, K.P.; Guan, C.; Tong, L.C.; Vinod, A.P. Discriminative FilterBank selection and EEG information fusion for Brain Computer Interface. In Proceedings of the 2009 IEEE International Symposium on Circuits and Systems, Taipei, Taiwan, 24–27 May 2009; pp. 1469–1472. [Google Scholar]
- Luo, J.; Feng, Z.; Lu, N. Spatio-temporal discrepancy feature for classification of motor imageries. Biomed. Signal Process. Control 2019, 47, 137–144. [Google Scholar] [CrossRef]
- Nicolas-Alonso, L.F.; Gomez-Gil, J. Brain Computer Interfaces, a Review. Sensors 2012, 12, 1211–1279. [Google Scholar] [CrossRef] [PubMed]
- Lotte, F. Signal Processing Approaches to Minimize or Suppress Calibration Time in Oscillatory Activity Based Brain Computer Interfaces. Proc. IEEE 2015, 103, 871–890. [Google Scholar] [CrossRef]
- Ang, K.K.; Chin, Z.Y.; Zhang, H.; Guan, C. Filter Bank Common Spatial Pattern (FBCSP) in Brain-Computer Interface. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 2390–2397. [Google Scholar]
- Lu, H.; Eng, H.; Guan, C.; Plataniotis, K.N.; Venetsanopoulos, A.N. Regularized Common Spatial Pattern with Aggregation for EEG Classification in Small-Sample Setting. IEEE Trans. Biomed. Eng. 2010, 57, 2936–2946. [Google Scholar]
- Dai, M.; Zheng, D.; Liu, S.; Zhang, P. Transfer Kernel Common Spatial Patterns for Motor Imagery Brain-Computer Interface Classification. Comput. Math. Methods Med. 2018. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Arvaneh, M.; Guan, C.; Ang, K.K.; Quek, H.C. Spatially sparsed Common Spatial Pattern to improve BCI performance. In Proceedings of the 2011 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Prague, Czech Republic, 22–27 May 2011; pp. 2412–2415. [Google Scholar]
- Lotte, F.; Guan, C. Spatially Regularized Common Spatial Patterns for EEG Classification. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 3712–3715. [Google Scholar]
- Park, Y.; Chung, W. BCI classification using locally generated CSP features. In Proceedings of the 2018 6th International Conference on Brain-Computer Interface (BCI), GangWon, South Korea, 15–17 January 2018; pp. 1–4. [Google Scholar]
- Yang, H.; Sakhavi, S.; Ang, K.K.; Guan, C. On the use of convolutional neural networks and augmented CSP features for multi-class motor imagery of EEG signals classification. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2620–2623. [Google Scholar]
- Park, S.; Lee, S. Small Sample Setting and Frequency Band Selection Problem Solving Using Subband Regularized Common Spatial Pattern. IEEE Sens. J. 2017, 17, 2977–2983. [Google Scholar] [CrossRef]
- Zhang, Y.; Nam, C.S.; Zhou, G.; Jin, J.; Wang, X.; Cichocki, A. Temporally Constrained Sparse Group Spatial Patterns for Motor Imagery BCI. IEEE Trans. Cybern. 2018. [Google Scholar] [CrossRef] [PubMed]
- Selim, S.; Tantawi, M.M.; Shedeed, H.A.; Badr, A. A CSP/AM—BA—SVM Approach for Motor Imagery BCI System. IEEE Access 2018, 6, 49192–49208. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Sun, S. Single-trial EEG classification of motor imagery using deep convolutional neural networks. Optik 2017, 130, 11–18. [Google Scholar] [CrossRef]
- Tabar, Y.R.; Halici, U. A novel deep learning approach for classification of EEG motor imagery signals. J. Neural Eng. 2017, 14, 016003. [Google Scholar] [CrossRef]
- Nguyen, C.H.; Artemiadis, P. EEG feature descriptors and discriminant analysis under Riemannian Manifold perspective. Neurocomputing 2018, 275, 1871–1883. [Google Scholar] [CrossRef]
- Barachant, A.; Bonnet, S.; Congedo, M.; Jutten, C. Multiclass Brain Computer Interface Classification by Riemannian Geometry. IEEE Trans. Biomed. Eng. 2012, 59, 920–928. [Google Scholar] [CrossRef]
- Davoudi, A.; Ghidary, S.S.; Sadatnejad, K. Dimensionality reduction based on distance preservation to local mean for symmetric positive definite matrices and its application in brain–computer interfaces. J. Neural Eng. 2017, 14, 036019. [Google Scholar] [CrossRef]
- Horev, I.; Yger, F.; Sugiyama, M. Geometry-aware principal component analysis for symmetric positive definite matrices. Mach. Learn. 2017, 106, 493–522. [Google Scholar] [CrossRef]
- Harandi, M.T.; Salzmann, M.; Hartley, R. From Manifold to Manifold: Geometry-Aware Dimensionality Reduction for SPD Matrices. In Computer Vision—ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 17–32. [Google Scholar]
- Kumar, S.; Mamun, K.; Sharma, A. CSP-TSM: Optimizing the performance of Riemannian tangent space mapping using common spatial pattern for MI-BCI. Comput. Biol. Med. 2017, 91, 231–242. [Google Scholar] [CrossRef]
- He, H.; Wu, D. Transfer Learning for Brain-Computer Interfaces: An Euclidean Space Data Alignment Approach. arXiv, 2018; arXiv:1808.05464. [Google Scholar]
- Yger, F.; Berar, M.; Lotte, F. Riemannian Approaches in Brain-Computer Interfaces: A Review. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Barachant, A.; Bonnet, S.; Congedo, M.; Jutten, C. Riemannian Geometry Applied to BCI Classification. In Latent Variable Analysis and Signal Separation; Vigneron, V., Zarzoso, V., Moreau, E., Gribonval, R., Vincent, E., Eds.; Springer: Berlin/Heidelberg, German, 2010; pp. 629–636. [Google Scholar]
- Li, X.; Wang, H. Smooth Spatial Filter for Common Spatial Patterns. In Neural Information Processing; Lee, M., Hirose, A., Hou, Z.G., Kil, R.M., Eds.; Springer: Berlin/Heidelberg, German, 2013; pp. 315–322. [Google Scholar]
- Blankertz, B.; Muller, K.; Krusienski, D.J.; Schalk, G.; Wolpaw, J.R.; Schlogl, A.; Pfurtscheller, G.; Millan, J.R.; Schroder, M.; Birbaumer, N. The BCI Competition III: Validating alternative approaches to actual BCI problems. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Tangermann, M.; Müller, K.R.; Aertsen, A.; Birbaumer, N.; Braun, C.; Brunner, C.; Leeb, R.; Mehring, C.; Miller, K.; Mueller-Putz, G.; et al. Review of the BCI Competition IV. Front. Neurosci. 2012, 6, 55. [Google Scholar]
- Arvaneh, M.; Guan, C.; Ang, K.K.; Quek, C. Optimizing the Channel Selection and Classification Accuracy in EEG-Based BCI. IEEE Trans. Biomed. Eng. 2011, 58, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Lotte, F.; Guan, C. Regularizing Common Spatial Patterns to Improve BCI Designs: Unified Theory and New Algorithms. IEEE Trans. Biomed. Eng. 2011, 58, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Pfurtscheller, G.; Neuper, C.; Flotzinger, D.; Pregenzer, M. EEG-based discrimination between imagination of right and left hand movement. Electroencephalogr. Clin. Neurophysiol. 1997, 103, 642–651. [Google Scholar] [CrossRef]
- Tang, Z.; Sun, S.; Zhang, S.; Chen, Y.; Li, C.; Chen, S. A Brain-Machine Interface Based on ERD/ERS for an Upper-Limb Exoskeleton Control. Sensors 2016, 16, 2050. [Google Scholar] [CrossRef]
- Belwafi, K.; Romain, O.; Gannouni, S.; Ghaffari, F.; Djemal, R.; Ouni, B. An embedded implementation based on adaptive filter bank for brain-computer interface systems. J. Neurosci. Methods 2018, 305, 1–16. [Google Scholar] [CrossRef]
- Selim, S.; Tantawi, M.; Shedeed, H.; Badr, A. Reducing Execution Time for Real-Time Motor Imagery Based BCI Systems. In Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2016; Hassanien, A.E., Shaalan, K., Gaber, T., Azar, A.T., Tolba, M.F., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 555–565. [Google Scholar]
- Gaur, P.; Pachori, R.B.; Wang, H.; Prasad, G. A multi-class EEG-based BCI classification using multivariate empirical mode decomposition based filtering and Riemannian geometry. Expert Syst. Appl. 2018, 95, 201–211. [Google Scholar] [CrossRef]
- Raza, H.; Cecotti, H.; Li, Y.; Prasad, G. Adaptive learning with covariate shift-detection for motor imagery-based brain–computer interface. Soft Comput. 2016, 20, 3085–3096. [Google Scholar] [CrossRef]



| BCI Competition (BCI-C) | BCI-C III | BCI-C IV | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | Dataset IVa | Dataset IIIa | Dataset IIa | ||||||||
| Electrodes | 118 | 60 | 22 | ||||||||
| Sampling Rate | 100 Hz | 250 Hz | 250 Hz | ||||||||
| Subject | aa | al | av | aw | ay | k3b | k6b | l1b | A01–A09 | ||
| Train | 168 | 224 | 84 | 56 | 28 | 90 | 60 | 60 | 144 | ||
| Test | 112 | 56 | 196 | 224 | 252 | 90 | 60 | 60 | 144 | ||
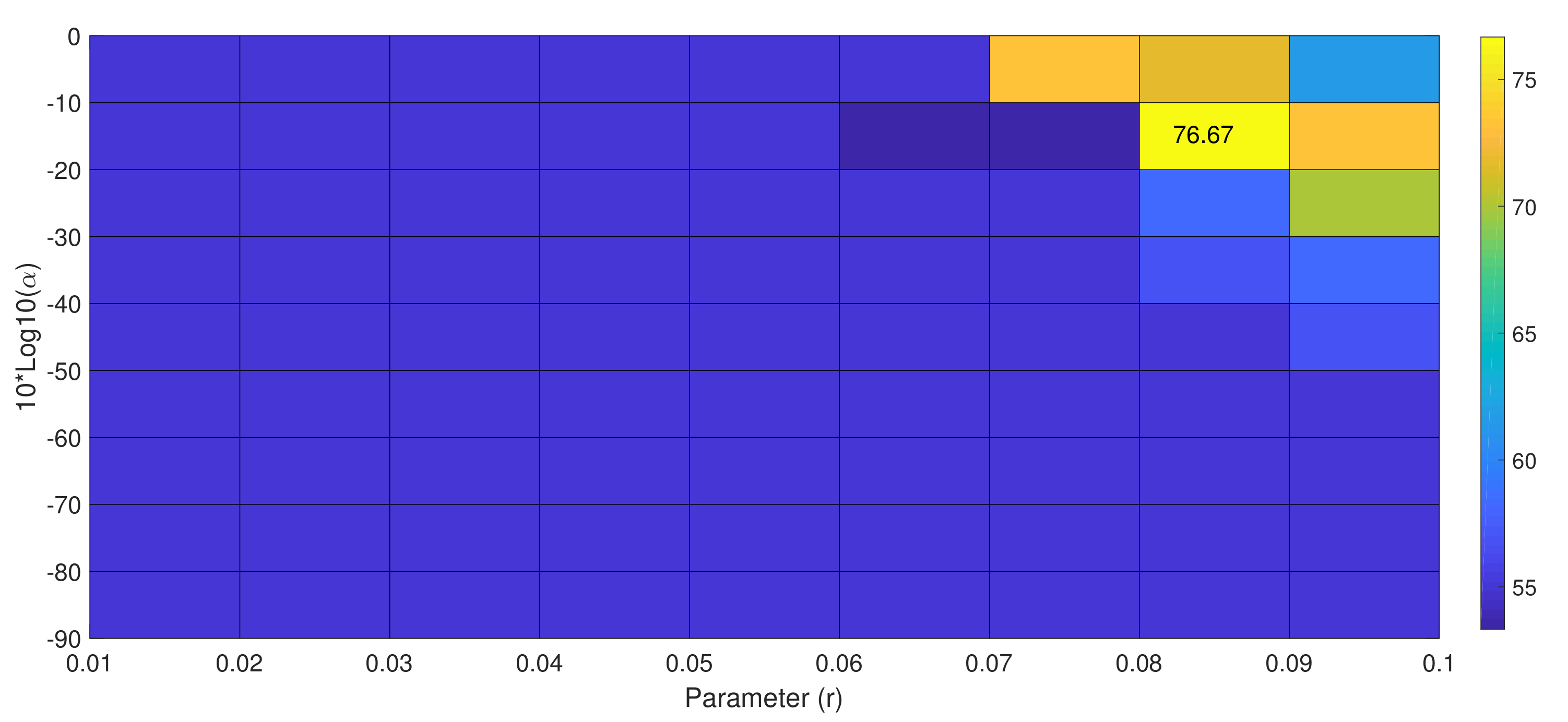
| Parameters | Dataset IVa | Dataset IIIa | Dataset IIa | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aa | al | av | aw | ay | k3b | k6b | l1b | A01 | A02 | A03 | A04 | A05 | A06 | A07 | A08 | A09 | |
| r | 0.06 | all r values | 0.07 | 0.07 | 0.08 | 0.1 | 0.08 | 0.01 | 0.06 | 0.07 | 0.05 | 0.09 | 0.07 | 0.04 | 0.06 | 0.01 | 0.01 |
| Studies | Methods | Year | aa | al | av | aw | ay | Mean | Std |
|---|---|---|---|---|---|---|---|---|---|
| Conventional Method | Csp | 66.07 | 96.43 | 47.45 | 71.88 | 49.6 | 66.28 | 19.83 | |
| Belwafi et al. [38] | Wola-Csp | 2018 | 66.07 | 96.07 | 52.14 | 71.43 | 50 | 67.29 | 18.54 |
| Arvaneh et al. [13] | Sscsp | 2011 | 72.32 | 96.42 | 54.10 | 70.53 | 73.41 | 73.35 | 15.09 |
| Lotte and Guan [14] | Srcsp | 2010 | 72.32 | 96.43 | 60.20 | 77.68 | 86.51 | 78.63 | 13.77 |
| Selim et al. [39] | Rms/Lda | 2016 | 69.64 | 89.29 | 59.18 | 88.84 | 86.90 | 78.77 | 13.65 |
| Dai et al. [11] | Tkcsp | 2018 | 68.10 | 93.88 | 68.47 | 88.40 | 74.93 | 79.17 | 11.78 |
| Park and Lee [17] | Sbrcsp | 2017 | 86.61 | 98.21 | 63.78 | 89.05 | 73.81 | 82.69 | 13.53 |
| Park and Chung [15] | Sss-Csp | 2018 | 74.11 | 100 | 67.78 | 90.07 | 89.29 | 84.46 | 13.05 |
| Selim et al. [19] | csp/am-ba-svm | 2018 | 86.61 | 100 | 66.84 | 90.63 | 80.95 | 85.00 | 12.30 |
| Proposed Method | sr-mdrm | 79.46 | 100 | 73.46 | 89.28 | 88.49 | 86.13 | 10.15 | |
| Wang et al. [32] | Winner | 96.00 | 100 | 81.00 | 100 | 98.00 | 94.20 | 8 |
| Studies | Year | aa | al | av | aw | ay | Mean |
|---|---|---|---|---|---|---|---|
| Barachant et al. [23]Mdrm | 0.22 | 0.86 | 0.25 | 0.13 | 0 | 0.29 | |
| Harandi et al. [26] Mdrm | 2014 | 0.23 | 1.00 | 0.40 | 0.53 | 0.82 | 0.59 |
| Horev et al. [25] Mdrm | 2017 | 0.62 | 0.96 | 0.42 | 0.68 | 0.60 | 0.65 |
| Davoudi et al. [24]-uDplm | 2017 | 0.57 | 1.00 | 0.39 | 0.64 | 0.72 | 0.66 |
| Davoudi et al. [24]-sDplm | 2017 | 0.63 | 1.00 | 0.46 | 0.66 | 0.78 | 0.70 |
| sr-Mdrm | 0.58 | 1 | 0.47 | 0.79 | 0.77 | 0.72 |
| Studies | Methods | Year | k3b | k6b | l1b | Mean | std |
|---|---|---|---|---|---|---|---|
| Proposed Method | sr-mdrm | 100 | 76.67 | 100 | 92.22 | 13.46 | |
| Zhang et al. [18] | Tsgsp | 2018 | 99.2 | 67.2 | 96.5 | 87.63 | 17.74 |
| Belwafi et al. [38] | wola-csp | 2018 | 97.77 | 61.66 | 93.33 | 84.25 | 19.69 |
| Conventional Method | csp | 95.56 | 61.67 | 93.33 | 83.52 | 18.95 | |
| Horev et al. [25] | horev-mdrm | 2017 | 95.56 | 68.33 | 85 | 82.96 | 13.72 |
| Barachant et al. [23] | mdrm | 96.66 | 60 | 88.33 | 81.66 | 19.21 | |
| Lotte and Guan [35] | srcsp | 2011 | 96.67 | 53.33 | 93.33 | 81.11 | 24.11 |
| Studies | Methods | Year | A01 | A02 | A03 | A04 | A05 | A06 | A07 | A08 | A09 | Mean | Std |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proposed Method | Sr-mdrm | 90.21 | 63.28 | 96.55 | 76.38 | 65.49 | 69.01 | 81.94 | 95.14 | 93.01 | 81.22 | 12.43 | |
| Gaur et al. [40] | ss-memdbf | 2018 | 91.49 | 60.56 | 94.16 | 76.16 | 58.52 | 68.52 | 78.57 | 97.01 | 93.85 | 79.93 | 14.14 |
| Barachant et al. [23] | Mdrm | 91.61 | 57.03 | 90.21 | 73.61 | 73.94 | 68.31 | 75 | 95.14 | 90.21 | 79.45 | 12.92 | |
| Belwafi et al. [38] | Wola-csp | 2018 | 86.81 | 63.19 | 94.44 | 68.75 | 56.25 | 69.44 | 78.47 | 97.91 | 93.75 | 78.85 | 15.15 |
| Lotte and Guan. [35] | srcsp | 2011 | 88.89 | 63.19 | 96.53 | 66.67 | 63.19 | 63.89 | 78.47 | 95.83 | 92.36 | 78.78 | 14.77 |
| standard Method | csp | 88.89 | 51.39 | 96.53 | 70.14 | 54.86 | 71.53 | 81.25 | 93.75 | 93.75 | 78.01 | 17.01 | |
| Raza et al. [41] | tlcsp | 2016 | 90.28 | 54.17 | 93.75 | 64.58 | 57.64 | 65.28 | 62.5 | 90.97 | 85.42 | 73.84 | 15.93 |
| Raza et al. [41] | tlcsp | 2016 | 90.28 | 57.64 | 95.14 | 65.97 | 61.11 | 65.28 | 61.11 | 91.67 | 86.11 | 74.92 | 15.42 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Lal, S.; Guesgen, H.W. Reduce Calibration Time in Motor Imagery Using Spatially Regularized Symmetric Positives-Definite Matrices Based Classification. Sensors 2019, 19, 379. https://doi.org/10.3390/s19020379
Singh A, Lal S, Guesgen HW. Reduce Calibration Time in Motor Imagery Using Spatially Regularized Symmetric Positives-Definite Matrices Based Classification. Sensors. 2019; 19(2):379. https://doi.org/10.3390/s19020379
Chicago/Turabian StyleSingh, Amardeep, Sunil Lal, and Hans W. Guesgen. 2019. "Reduce Calibration Time in Motor Imagery Using Spatially Regularized Symmetric Positives-Definite Matrices Based Classification" Sensors 19, no. 2: 379. https://doi.org/10.3390/s19020379
APA StyleSingh, A., Lal, S., & Guesgen, H. W. (2019). Reduce Calibration Time in Motor Imagery Using Spatially Regularized Symmetric Positives-Definite Matrices Based Classification. Sensors, 19(2), 379. https://doi.org/10.3390/s19020379





