Spectral Imaging Analysis for Ultrasensitive Biomolecular Detection Using Gold-Capped Nanowire Arrays
Abstract
:1. Introduction
2. Experimental Section
2.1. Chip Fabrication
2.2. Optical Setup
3. Result and Discussion
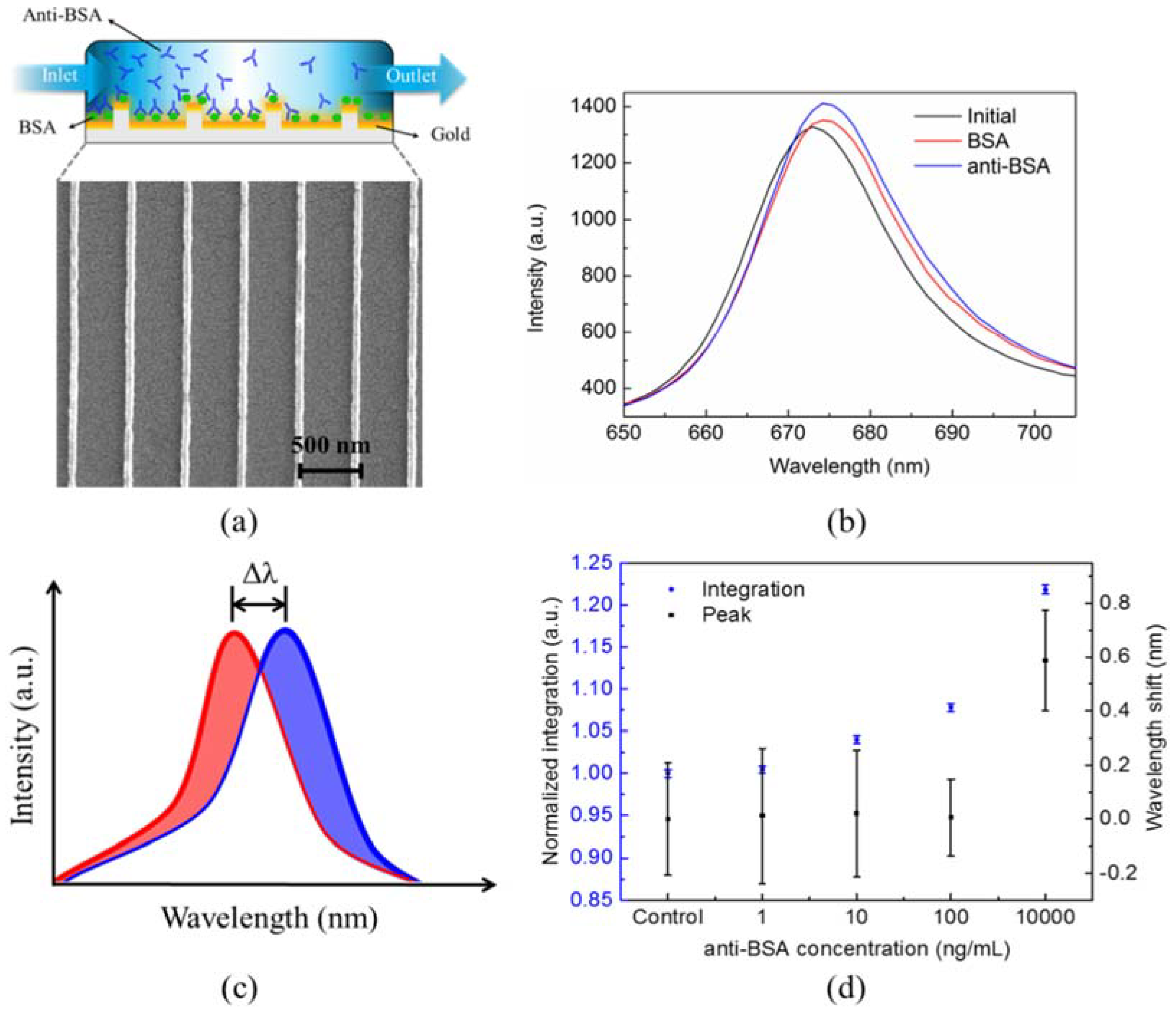
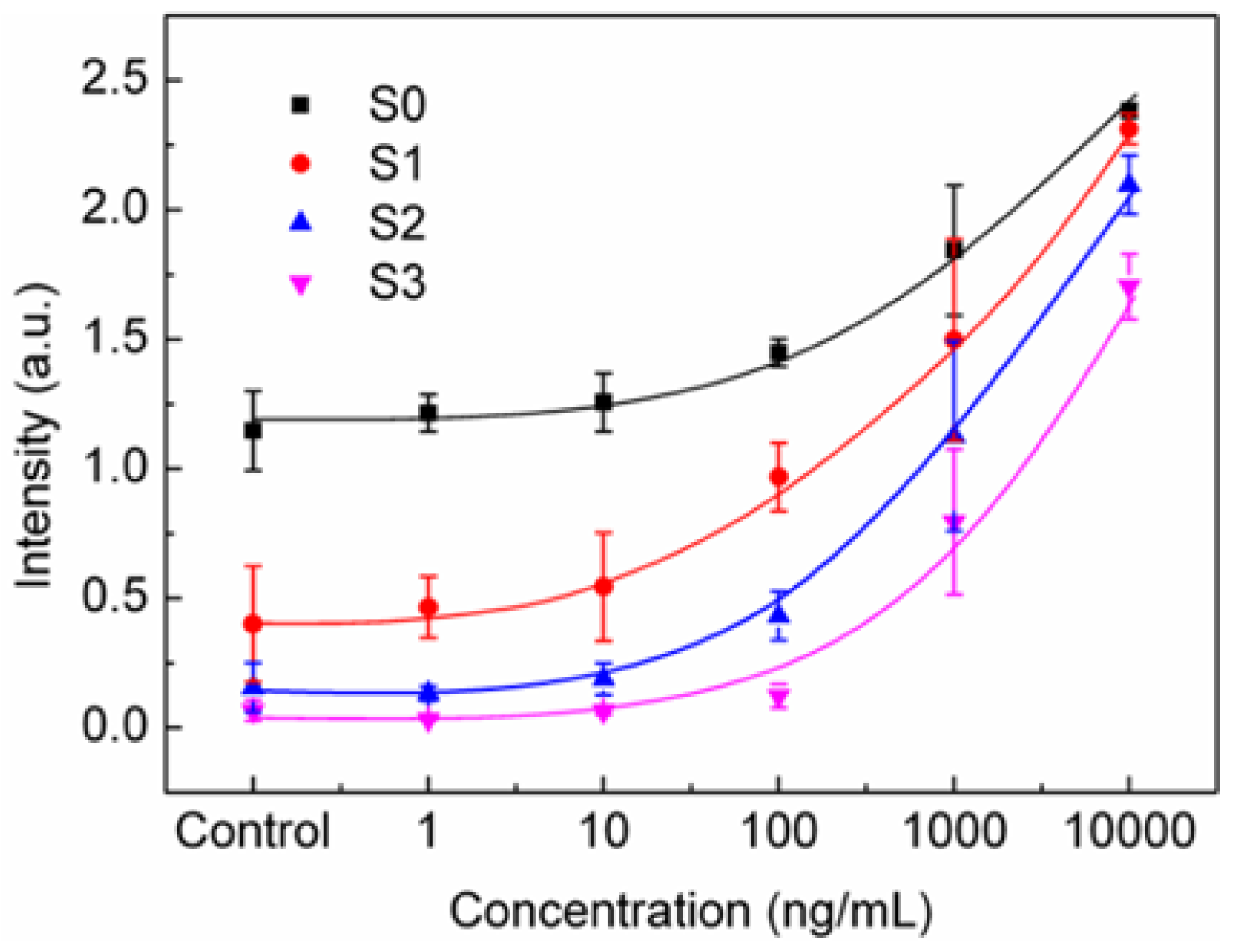
3.1. Spectral Integration Analysis (SIA)
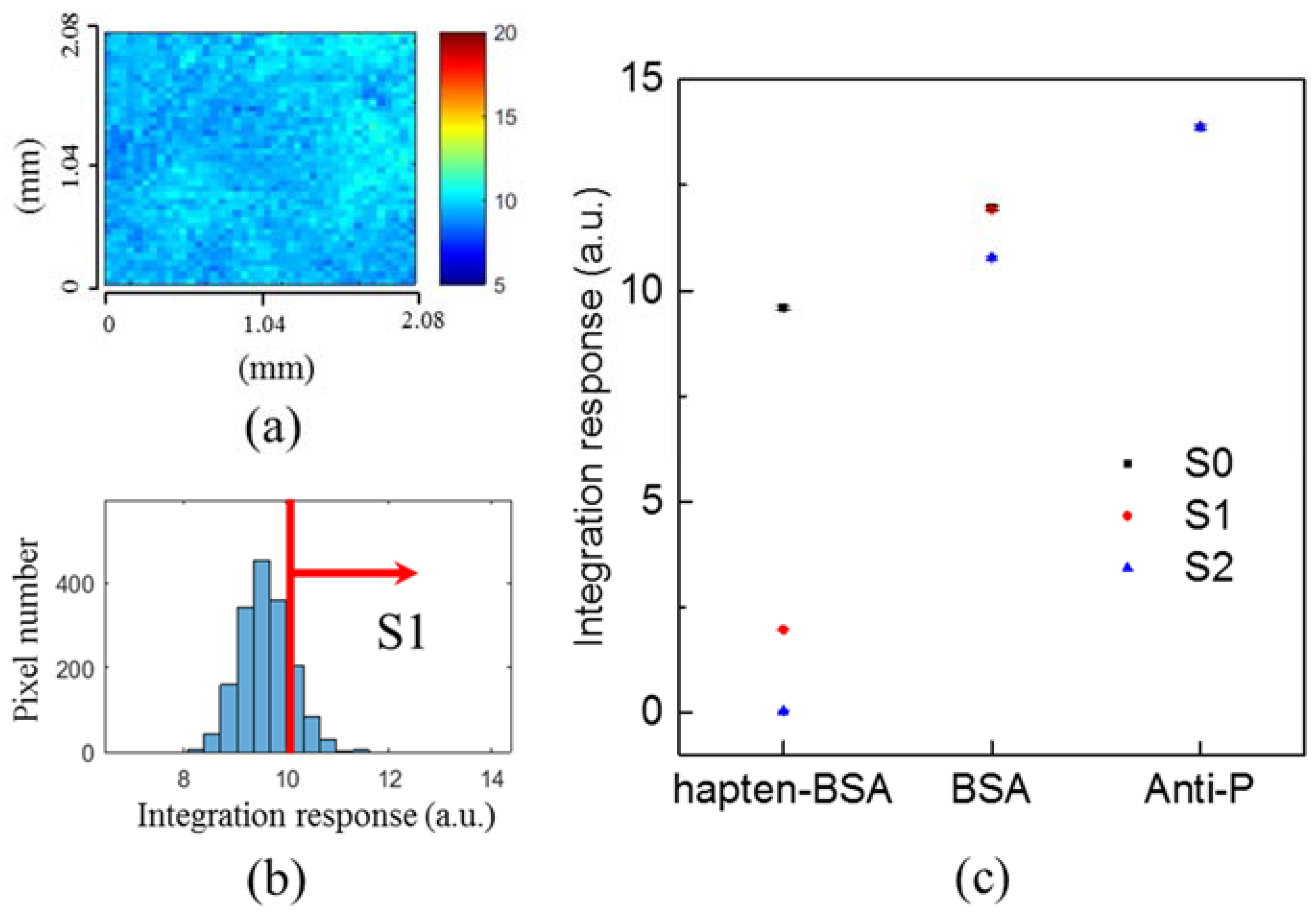
3.2. Threshold Method
3.3. Experiments in Phosphate-Buffered Saline (PBS)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jordan, C.E.; Frutos, A.G.; Thiel, A.J.; Corn, R.M. Surface plasmon resonance imaging measurements of DNA hybridization adsorption and streptavidin/DNA multilayer formation at chemically modified gold surfaces. Anal. Chem. 1997, 69, 4939–4947. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, L.; Wu, S.Y.; He, J.; Qu, J.; Li, X.; Ho, H.P.; Gu, D.; Gao, B.Z.; Shao, Y. Wavelength-Scanning SPR Imaging Sensors Based on an Acousto-Optic Tunable Filter and a White Light Laser. Sensors 2017, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- Brockman, J.M.; Frutos, A.G.; Corn, R.M. A multistep chemical modification procedure to create DNA arrays on gold surfaces for the study of protein-DNA interactions with surface plasmon resonance imaging. J. Am. Chem. Soc. 1999, 121, 8044–8051. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, L.; Wu, S.Y.; He, J.; Qu, J.; Li, X.; Ho, H.P.; Gu, D.; Gao, B.Z.; Shao, Y. High-throughput imaging surface plasmon resonance biosensing based on an adaptive spectral-dip tracking scheme. Opt. Express 2016, 24, 28303–28311. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, L.; Liu, L.; Chong, X.; Guo, J.; Ma, S.; Ji, Y.; He, Y. Parallel-scan based microarray imager capable of simultaneous surface plasmon resonance and hyperspectral fluorescence imaging. Biosens. Bioelectron. 2011, 30, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.P.; Frutos, A.G.; Brockman, J.M.; Corn, R.M. Near-infrared surface plasmon resonance measurements of ultrathin films. 1. Angle shift and SPR imaging experiments. Anal. Chem. 1999, 71, 3928–3934. [Google Scholar] [CrossRef]
- Nelson, S.G.; Johnston, K.S.; Yee, S.S. High sensitivity surface plasmon resonance sensor based on phase detection. Sens. Actuator B Chem. 1996, 35, 187–191. [Google Scholar] [CrossRef]
- Wong, C.L.; Ho, H.P.; Yu, T.T.; Suen, Y.K.; Chow, W.W.Y.; Wu, S.Y.; Law, W.C.; Yuan, W.; Li, W.J.; Kong, S.K.; et al. Two-dimensional biosensor arrays based on surface plasmon resonance phase imaging. Appl. Opt. 2007, 46, 2325–2332. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.L.; Ho, H.P.; Suen, Y.K.; Kong, S.K.; Chen, Q.L.; Yuan, W.; Wu, S.Y. Real-time protein biosensor arrays based on surface plasmon resonance differential phase imaging. Biosens. Bioelectron. 2008, 24, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lindquist, N.C.; Wittenberg, N.J.; Jordan, L.R.; Oh, S.H. Real-time full-spectral imaging and affinity measurements from 50 microfluidic channels using nanohole surface plasmon resonance. Lab Chip 2012, 12, 3882–3890. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.S.; Kester, R.T.; Lewis, P.E.; Bedard, N.; Tkaczyk, T.S. Image mapping spectrometry: A novel hyperspectral platform for rapid snapshot imaging. Int. Soc. Opt. Photonics 2011, 8048, 80480J. [Google Scholar]
- Wood, J.B.; Szyndler, M.W.; Halpern, A.R.; Cho, K.; Corn, R.M. Fabrication of DNA microarrays on polydopamine-modified gold thin films for SPR imaging measurements. Langmuir 2013, 29, 10868–10873. [Google Scholar] [CrossRef] [PubMed]
- Premaratne, G.; Mubarak, Z.H.A.; Senavirathna, L.; Liu, L.; Krishnan, S. Measuring ultra-low levels of nucleotide biomarkers using quartz crystal microbalance and SPR microarray imaging methods: A comparative analysis. Sens. Actuators B Chem. 2017, 253, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Chong, X.; Liu, L.; Liu, Z.; Ma, S.; Guo, J.; Ji, Y.; He, Y. Detect the Hybridization of Single-Stranded DNA by Parallel Scan Spectral Surface Plasmon Resonance Imaging. Plasmonics 2013, 8, 1185–1191. [Google Scholar] [CrossRef]
- Hänggi, P.; Marchesoni, F. Artificial Brownian motors: Controlling transport on the nanoscale. Rev. Mod. Phys. 2009, 81, 387–442. [Google Scholar] [CrossRef] [Green Version]
- Astumian, R.D. Thermodynamics and kinetics of a Brownian motor. Science 1997, 276, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Adrian, R.J. Particle-imaging techniques for experimental fluid-mechanics. Annu. Rev. Fluid Mech. 1991, 23, 261–304. [Google Scholar] [CrossRef]
- Stickel, J.J.; Powell, R.L. Fluid Mechanics and Rheology of Dense Suspensions. Annu. Rev. Fluid Mech. 2005, 37, 129–149. [Google Scholar] [CrossRef]
- Mandracchia, B.; Pagliarulo, V.; Paturzo, M.; Ferraro, P. Surface plasmon resonance imaging by holographic enhanced mapping. Anal. Chem. 2015, 87, 4124–4128. [Google Scholar] [CrossRef] [PubMed]
- Malic, L.; Veres, T.; Tabrizian, M. Nanostructured digital microfluidics for enhanced surface plasmon resonance imaging. Biosens. Bioelectron. 2011, 26, 2053–2059. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Huang, J.B.; Chang, J.W.; Wu, S.H.; Wei, P.K. Ultrasensitive biosensors using enhanced Fano resonances in capped gold nanoslit arrays. Sci. Rep. 2015, 5, 8547. [Google Scholar] [CrossRef] [PubMed]
- Reiner, A.T.; Ferrer, N.G.; Venugopalan, P.; Lai, R.C.; Lim, S.K.; Dostalek, J. Magnetic nanoparticle-enhanced surface plasmon resonance biosensor for extracellular vesicle analysis. Analyst 2017, 142, 3913–3921. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Peng, Y.; Bai, J.; Zhang, X.; Cui, Y.; Ning, B.; Cui, J.; Gao, Z. Magnetic nanoparticle enhanced surface plasmon resonance sensor for estradiol analysis. Sens. Actuators B Chem. 2018, 254, 629–635. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, Y.; Zhang, D.; Li, S.; Zhang, Y.; Ma, P.; Yu, Y.; Wang, X.; Song, D. Ultrasensitive magnetic field-assisted surface plasmon resonance immunoassay for human cardiac troponin I. Biosens. Bioelectron. 2017, 96, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Barik, A.; Otto, L.M.; Yoo, D.; Jose, J.; Johnson, T.W.; Oh, S.H. Dielectrophoresis-enhanced plasmonic sensing with gold nanohole arrays. Nano Lett. 2014, 14, 2006–2012. [Google Scholar] [CrossRef] [PubMed]
- Kyaw, H.H.; Boonruang, S.; Mohammed, W.S.; Dutta, J. Design of electric-field assisted surface plasmon resonance system for the detection of heavy metal ions in water. AIP Adv. 2015, 5, 107226. [Google Scholar] [CrossRef] [Green Version]
- Malic, L.; Veres, T.; Tabrizian, M. Biochip functionalization using electrowetting-on-dielectric digital microfluidics for surface plasmon resonance imaging detection of DNA hybridization. Biosens. Bioelectron. 2009, 24, 2218–2224. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Wu, T.Y.; Hsu, H.Y.; Yang, S.Y.; Wei, P.K. Low-Cost and Rapid Fabrication of Metallic Nanostructures for Sensitive Biosensors Using Hot-Embossing and Dielectric-Heating Nanoimprint Methods. Sensors 2017, 17, 1548. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Hsu, H.Y.; You, M.L.; Chang, C.C.; Pan, M.Y.; Shi, X.; Ueno, K.; Misawa, H.; Wei, P.K. Highly Sensitive Aluminum-Based Biosensors using Tailorable Fano Resonances in Capped Nanostructures. Sci. Rep. 2017, 7, 44104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, E.H.; Tsai, W.S.; Lee, K.L.; Lee, M.C.; Wei, P.K. Enhancing detection sensitivity of metallic nanostructures by resonant coupling mode and spectral integration analysis. Opt. Express 2014, 22, 19621–19632. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Chih, M.J.; Shi, X.; Ueno, K.; Misawa, H.; Wei, P.K. Improving surface plasmon detection in gold nanostructures using a multi-polarization spectral integration method. Adv. Mater. 2012, 24, OP253–OP259. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tai, Y.-H.; Fu, P.-H.; Lee, K.-L.; Wei, P.-K. Spectral Imaging Analysis for Ultrasensitive Biomolecular Detection Using Gold-Capped Nanowire Arrays. Sensors 2018, 18, 2181. https://doi.org/10.3390/s18072181
Tai Y-H, Fu P-H, Lee K-L, Wei P-K. Spectral Imaging Analysis for Ultrasensitive Biomolecular Detection Using Gold-Capped Nanowire Arrays. Sensors. 2018; 18(7):2181. https://doi.org/10.3390/s18072181
Chicago/Turabian StyleTai, Yi-Hsin, Po-Han Fu, Kuang-Li Lee, and Pei-Kuen Wei. 2018. "Spectral Imaging Analysis for Ultrasensitive Biomolecular Detection Using Gold-Capped Nanowire Arrays" Sensors 18, no. 7: 2181. https://doi.org/10.3390/s18072181




