Feature Fusion of ICP-AES, UV-Vis and FT-MIR for Origin Traceability of Boletus edulis Mushrooms in Combination with Chemometrics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Instruments and Reagents
2.3. Data Analysis
2.4. Software
3. Results and Discussion
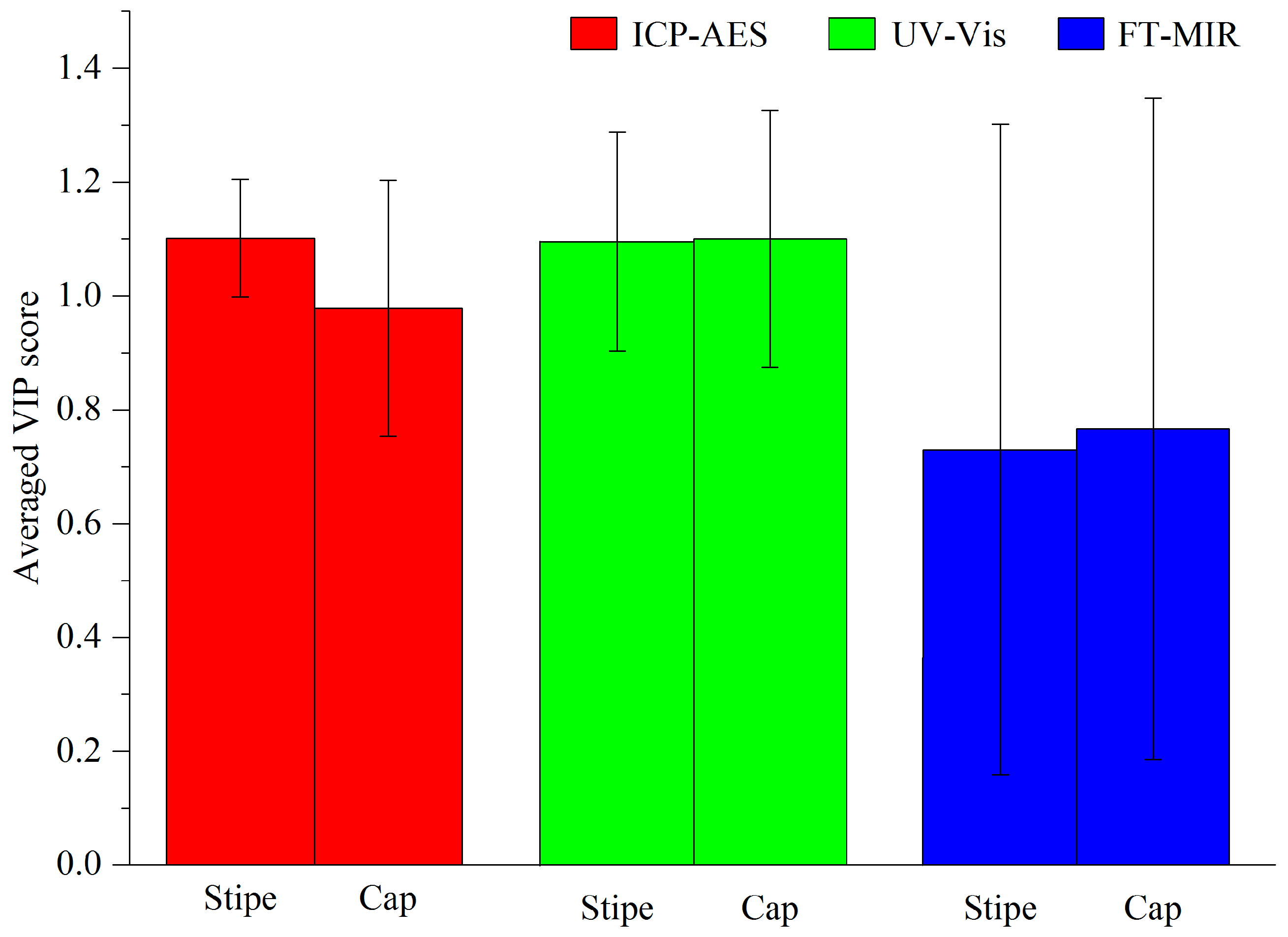
3.1. Comparison Analysis between Cap and Stipe
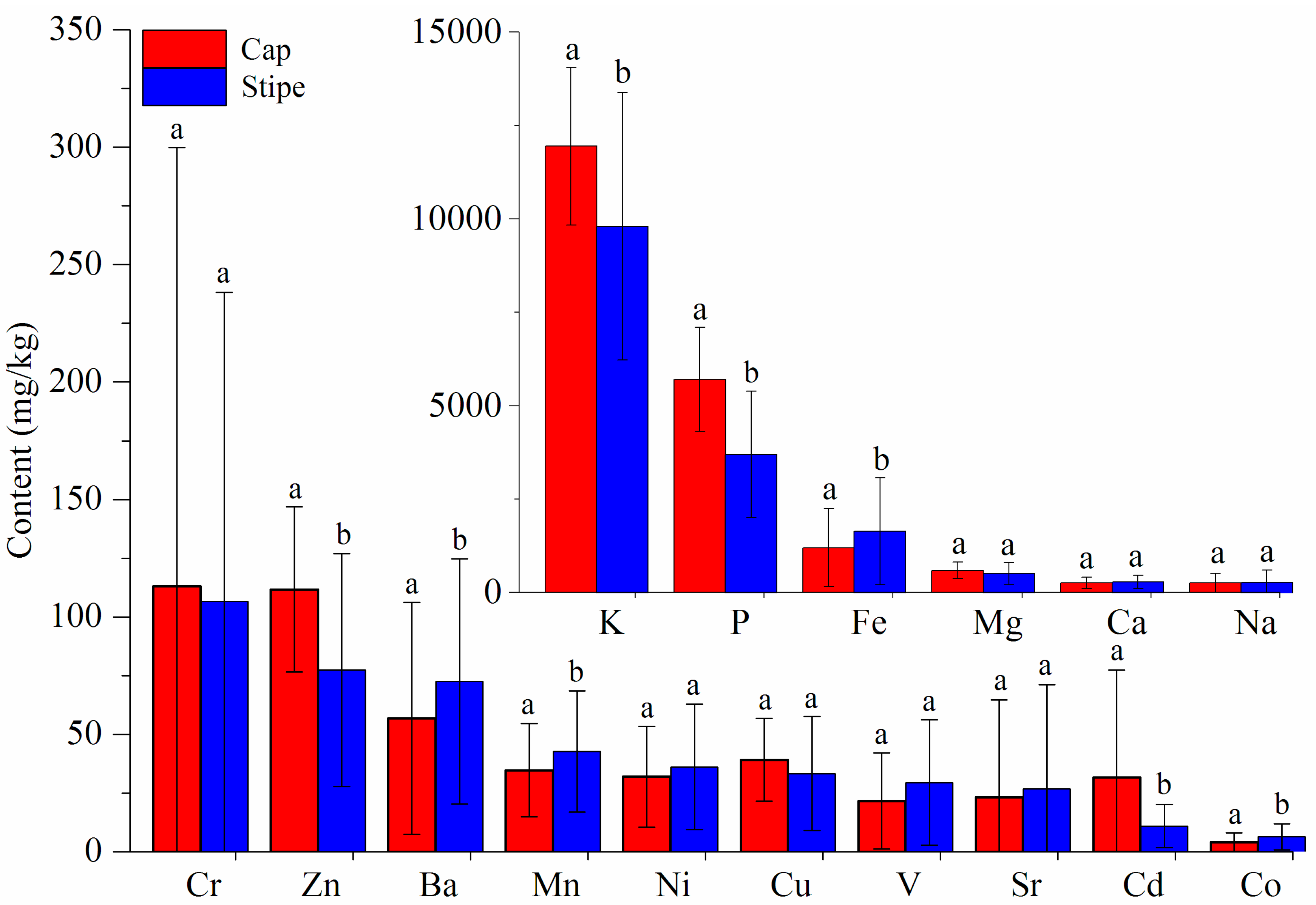
3.1.1. ICP-AES
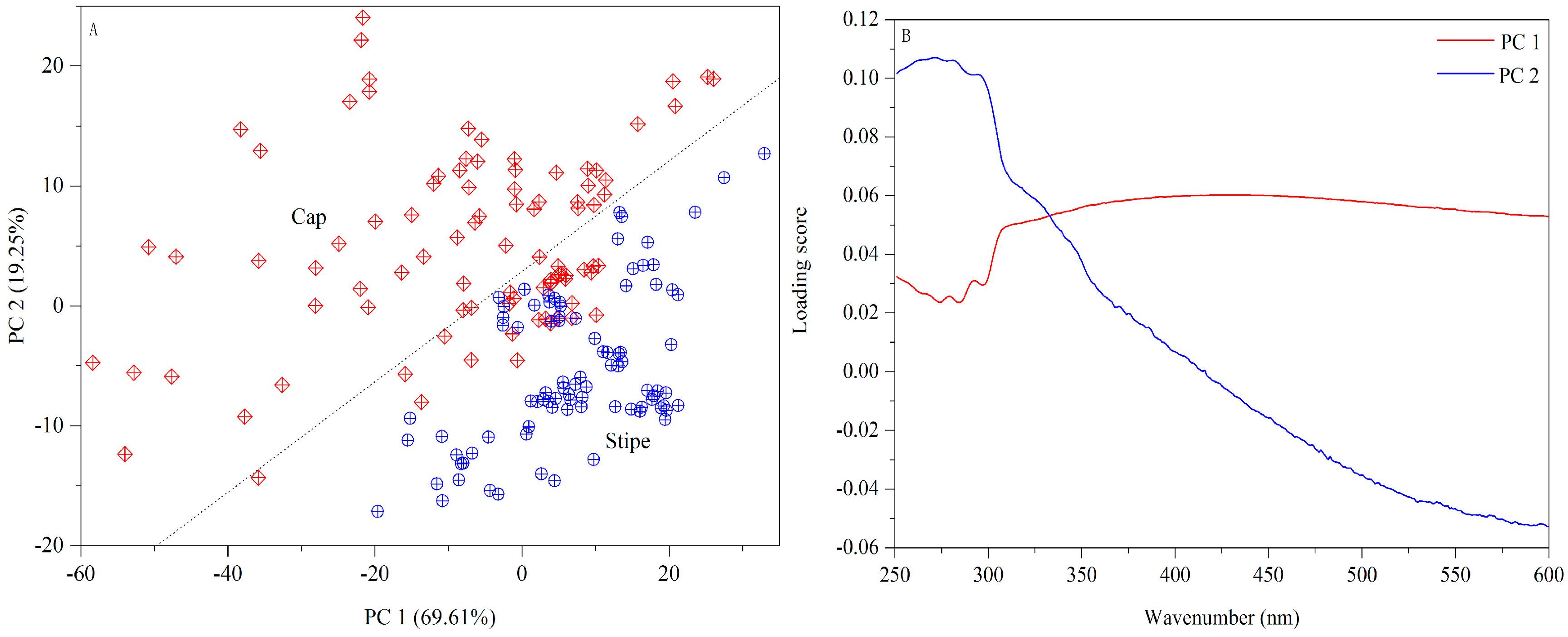
3.1.2. UV-Vis
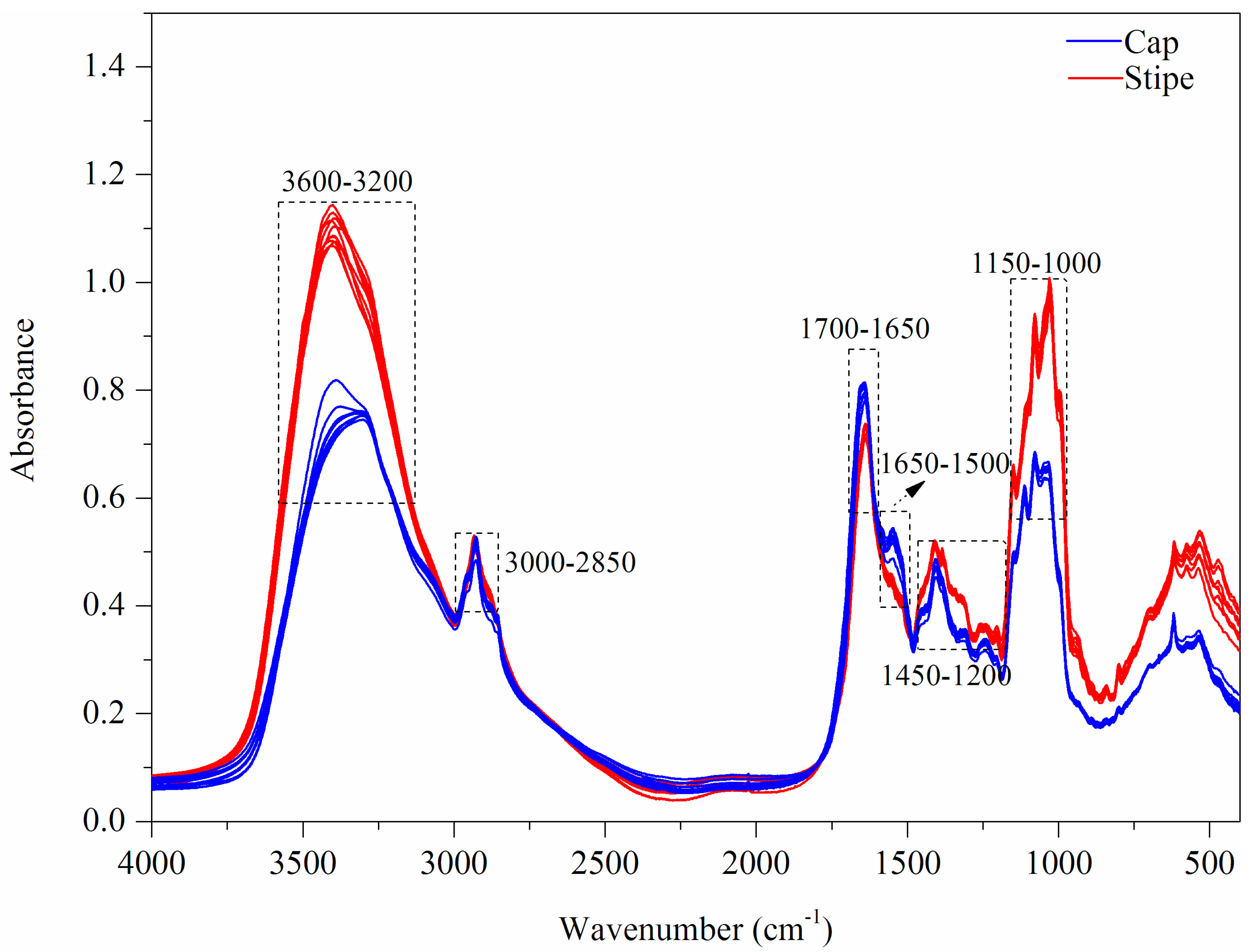
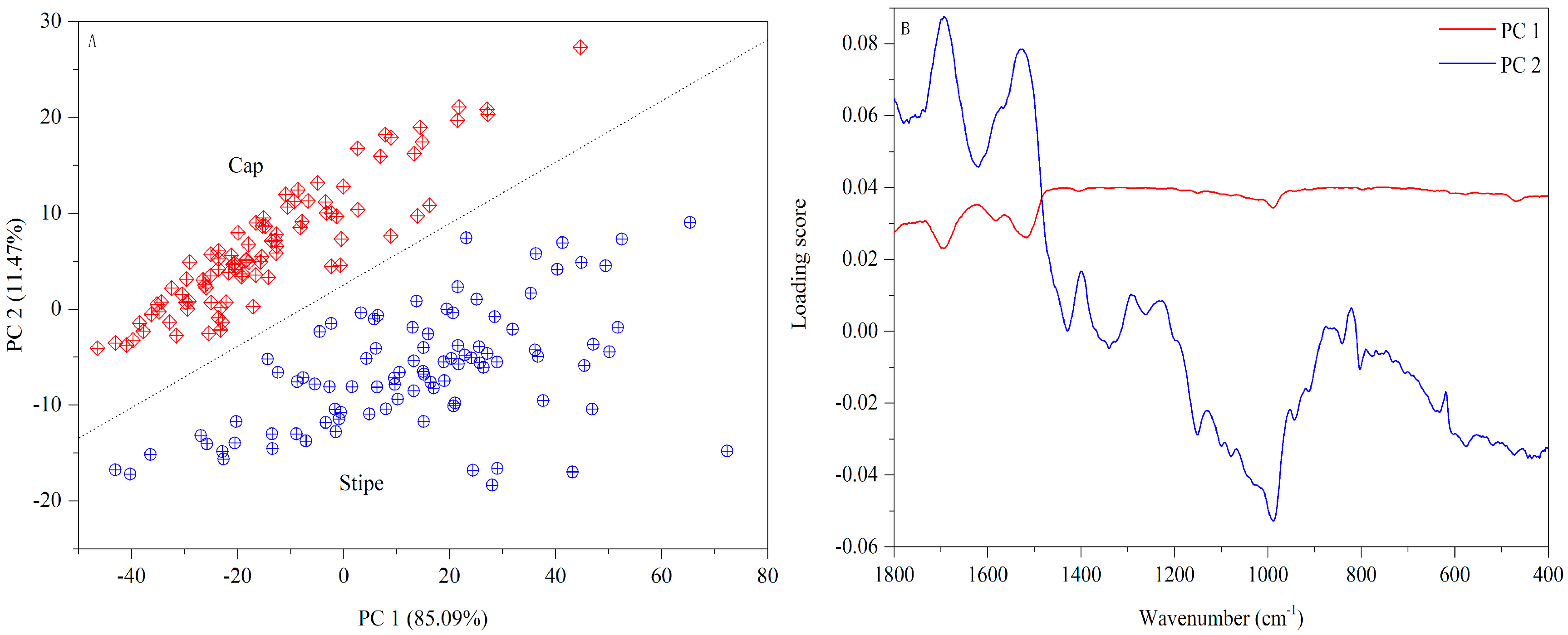
3.1.3. FT-MIR
3.2. Regional Differences Based on VIP
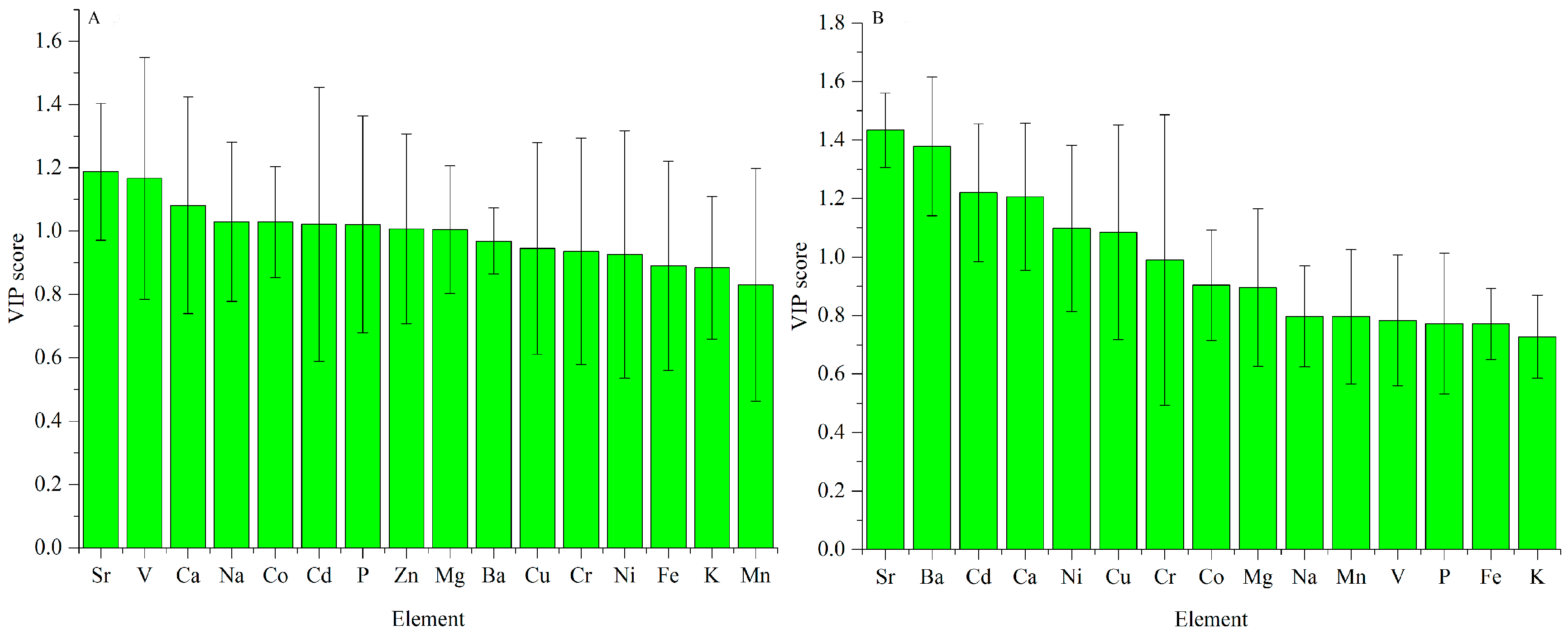
3.2.1. ICP-AES Dataset
3.2.2. UV-Vis Dataset
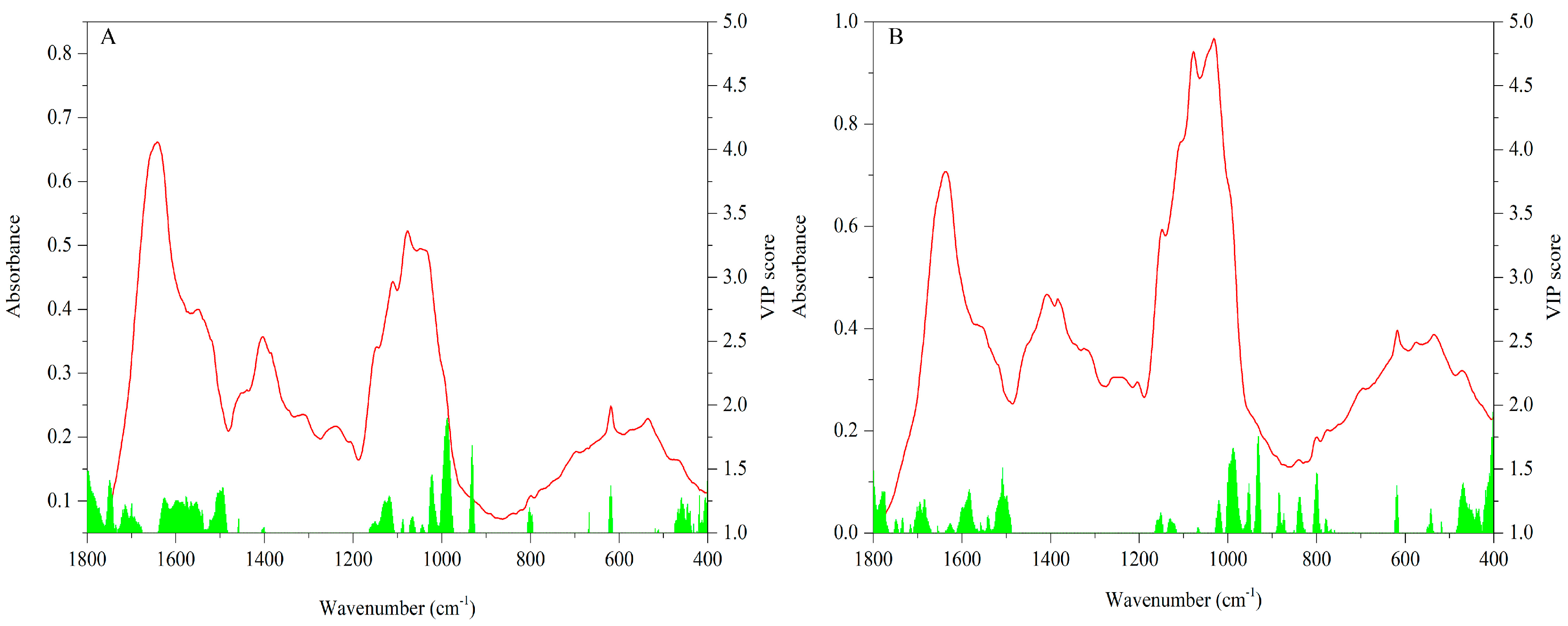
3.2.3. FT-MIR Dataset
3.3. Origin Traceability Based on Chemometrics
3.3.1. Cap
3.3.2. Stipe
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fernandes, Â.; Petrović, J.; Stojković, D.; Barros, L.; Glamočlija, J.; Soković, M.; Martins, A.; Ferreira, I.C.F.R. Polyporus squamosus (Huds.) Fr from different origins: Chemical characterization, screening of the bioactive properties and specific antimicrobial effects against Pseudomonas aeruginosa. LWT Food Sci. Technol. 2016, 69, 91–97. [Google Scholar] [CrossRef]
- Tesanovic, K.; Pejin, B.; Sibul, F.; Matavulj, M.; Raseta, M.; Janjusevic, L.; Karaman, M. A comparative overview of antioxidative properties and phenolic profiles of different fungal origins: Fruiting bodies and submerged cultures of Coprinus comatus and Coprinellus truncorum. J. Food Sci. Technol. 2017, 54, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Li, T.; Liu, H.; Li, J.; Wang, Y. Traceability of Boletaceae mushrooms using data fusion of UV-visible and FTIR combined with chemometrics methods. J. Sci. Food Agric. 2017. [Google Scholar] [CrossRef] [PubMed]
- Falandysz, J.; Frankowska, A.; Mazur, A. Mercury and its bioconcentration factors in King Bolete (Boletus edulis) Bull. Fr. J. Environ. Sci. Health Part A 2007, 42, 2089–2095. [Google Scholar] [CrossRef] [PubMed]
- Falandysz, J.; Zhang, J.; Wang, Y.Z.; Saba, M.; Krasinska, G.; Wiejak, A.; Li, T. Evaluation of Mercury Contamination in Fungi Boletus Species from Latosols, Lateritic Red Earths, and Red and Yellow Earths in the Circum-Pacific Mercuriferous Belt of Southwestern China. PLoS ONE 2015, 10, e0143608. [Google Scholar] [CrossRef] [PubMed]
- Palacios, I.; Lozano, M.; Moro, C.; D’Arrigo, M.; Rostagno, M.A.; Martínez, J.A.; García-Lafuente, A.; Guillamón, E.; Villares, A. Antioxidant properties of phenolic compounds occurring in edible mushrooms. Food Chem. 2011, 128, 674–678. [Google Scholar] [CrossRef]
- Tsai, S.Y.; Tsai, H.L.; Mau, J.L. Antioxidant properties of Agaricus blazei, Agrocybe cylindracea, and Boletus edulis. LWT Food Sci. Technol. 2007, 40, 1392–1402. [Google Scholar] [CrossRef]
- Sarikurkcu, C.; Tepe, B.; Yamac, M. Evaluation of the antioxidant activity of four edible mushrooms from the Central Anatolia, Eskisehir-Turkey: Lactarius deterrimus, Suillus collitinus, Boletus edulis, Xerocomus chrysenteron. Bioresour. Technol. 2008, 99, 6651–6655. [Google Scholar] [CrossRef] [PubMed]
- Sitta, N.; Floriani, M. Nationalization and globalization trends in the wild mushroom commerce of Italy with emphasis on porcini (Boletus edulis and allied species). Econ. Bot. 2008, 62, 307–322. [Google Scholar] [CrossRef]
- Ulziijargal, E.; Mau, J.L. Nutrient compositions of culinary-medicinal mushroom fruiting bodies and mycelia. Int. J. Med. Mushrooms 2011, 13, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Xiao, N.; He, P.; Sun, P. Chemical analysis and antioxidant activity in vitro of polysaccharides extracted from Boletus edulis. Int. J. Biol. Macromol. 2011, 49, 1092–1095. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, B.; Lopes, R.; Andrade, P.B.; Seabra, R.M.; Goncalves, R.F.; Baptista, P.; Quelhas, I.; Valentao, P.C. Comparative study of phytochemicals and antioxidant potential of wild edible mushroom caps and stipes. Food Chem. 2008, 110, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Gachumi, G.; El-Aneed, A. Mass Spectrometric Approaches for the Analysis of Phytosterols in Biological Samples. J. Agric. Food Chem. 2017, 65, 10141–10156. [Google Scholar] [CrossRef] [PubMed]
- Donno, D.; Beccaro, G.L.; Mellano, M.G.; Cerutti, A.K.; Canterino, S.; Bounous, G. Effect of agronomic and environmental conditions on chemical composition of tree-species buds used for herbal preparations. Vegetos Int. J. Plant Res. 2012, 25, 21–29. [Google Scholar]
- Qi, L.M.; Zhang, J.; Liu, H.G.; Li, T.; Wang, Y.Z. Fourier transform mid-infrared spectroscopy and chemometrics to identify and discriminate Boletus edulis and Boletus tomentipes mushrooms. Int. J. Food Prop. 2017, 20. [Google Scholar] [CrossRef]
- Botelho, B.G.; Oliveira, L.S.; Franca, A.S. Fluorescence spectroscopy as tool for the geographical discrimination of coffees produced in different regions of Minas Gerais State in Brazil. Food Control 2017, 77, 25–31. [Google Scholar] [CrossRef]
- Jiang, W.; Xie, C.; Zhuang, M.; Shou, Y.; Tang, Y. Sensor Data Fusion with Z-Numbers and Its Application in Fault Diagnosis. Sensors 2016, 16, 1509. [Google Scholar] [CrossRef] [PubMed]
- Biancolillo, A.; Bucci, R.; Magri, A.L.; Magri, A.D.; Marini, F. Data-fusion for multiplatform characterization of an Italian craft beer aimed at its authentication. Anal. Chim. Acta 2014, 820, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Alamprese, C.; Casale, M.; Sinelli, N.; Lanteri, S.; Casiraghi, E. Detection of minced beef adulteration with turkey meat by UV-Vis, NIR and MIR spectroscopy. LWT Food Sci. Technol. 2013, 53, 225–232. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, X.; Zhang, Z.; Zhu, R. Data fusion of near-infrared and mid-infrared spectra for identification of rhubarb. Spectrochim. Acta Part A 2017, 171, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Li, T.; Yang, T.; Wang, Y.; Liu, H. Ultraviolet Spectroscopy Used to Fingerprint Five Wild-Grown Edible Mushrooms (Boletaceae) Collected from Yunnan, China. J. Spectrosc. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Donno, D.; Boggia, R.; Zunin, P.; Cerutti, A.K.; Guido, M.; Mellano, M.G.; Prgome, Z.; Beccaro, G.L. Phytochemical fingerprint and chemometrics for natural food preparation pattern recognition: An innovative technique in food supplement quality control. J. Food Sci. Technol. 2016, 53, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Peev, C.I.; Vlase, L.; Antal, D.S.; Dehelean, C.A.; Szabadai, Z. Determination of some polyphenolic compounds in buds of Alnus and Corylus species by HPLC. Chem. Nat. Compd. 2007, 43, 259–262. [Google Scholar] [CrossRef]
- Lim, J.; Kim, G.; Mo, C.; Oh, K.; Yoo, H.; Ham, H.; Kim, M.S. Classification of Fusarium-Infected Korean Hulled Barley Using Near-Infrared Reflectance Spectroscopy and Partial Least Squares Discriminant Analysis. Sensors 2017, 17, 2258. [Google Scholar] [CrossRef] [PubMed]
- Galtier, O.; Abbas, O.; Le Dréau, Y.; Rebufa, C.; Kister, J.; Artaud, J.; Dupuy, N. Comparison of PLS1-DA, PLS2-DA and SIMCA for classification by origin of crude petroleum oils by MIR and virgin olive oils by NIR for different spectral regions. Vib. Spectrosc. 2011, 55, 132–140. [Google Scholar] [CrossRef]
- Hirri, A.; Bassbasi, M.; Platikanov, S.; Tauler, R.; Oussama, A. FTIR Spectroscopy and PLS-DA Classification and Prediction of Four Commercial Grade Virgin Olive Oils from Morocco. Food Anal. Methods 2015, 9, 974–981. [Google Scholar] [CrossRef]
- Dupuy, N.; Galtier, O.; Ollivier, D.; Vanloot, P.; Artaud, J. Comparison between NIR, MIR, concatenated NIR and MIR analysis and hierarchical PLS model. Application to virgin olive oil analysis. Anal. Chim. Acta 2010, 666, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.-M.; Zhang, J.; Zuo, Z.-T.; Zhao, Y.-L.; Wang, Y.-Z.; Hang, J. Determination of Iridoids in Gentiana rigescens by Infrared Spectroscopy and Multivariate Analysis. Anal. Lett. 2016, 50, 389–401. [Google Scholar] [CrossRef]
- Mahadevan, S.; Shah, S.L.; Marrie, T.J.; Slupsky, C.M. Analysis of metabolomic data using support vector machines. Anal. Chem. 2008, 80, 7562–7570. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Wang, J. Detection of adulteration in cherry tomato juices based on electronic nose and tongue: Comparison of different data fusion approaches. J. Food Eng. 2014, 126, 89–97. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Heberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhao, J.; Fang, C.H.; Wang, D. Feasibility study on identification of green, black and Oolong teas using near-infrared reflectance spectroscopy based on support vector machine (SVM). Spectrochim. Acta Part A 2007, 66, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Nikkarinen, M.; Mertanen, E. Impact of geological origin on trace element composition of edible mushrooms. J. Food Compos. Anal. 2004, 17, 301–310. [Google Scholar] [CrossRef]
- Jarzyńska, G.; Falandysz, J. Trace elements profile of Slate Bolete (Leccinum duriusculum) mushroom and associated upper soil horizon. J. Geochem. Explor. 2012, 121, 69–75. [Google Scholar] [CrossRef]
- Giannaccini, G.; Betti, L.; Palego, L.; Mascia, G.; Schmid, L.; Lanza, M.; Mela, A.; Fabbrini, L.; Biondi, L.; Lucacchini, A. The trace element content of top-soil and wild edible mushroom samples collected in Tuscany, Italy. Environ. Monit. Assess. 2012, 184, 7579–7595. [Google Scholar] [CrossRef] [PubMed]
- Chudzynski, K.; Falandysz, J. Multivariate analysis of elements content of Larch Bolete (Suillus grevillei) mushroom. Chemosphere 2008, 73, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Anghileri, A.; Lantto, R.; Kruus, K.; Arosio, C.; Freddi, G. Tyrosinase-catalyzed grafting of sericin peptides onto chitosan and production of protein-polysaccharide bioconjugates. J. Biotechnol. 2007, 127, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Gonzaga, M.L.C.; Ricardo, N.M.P.S.; Heatley, F.; Soares, S.d.A. Isolation and characterization of polysaccharides from Agaricus blazei Murill. Carbohydr. Polym. 2005, 60, 43–49. [Google Scholar] [CrossRef]
- Hung, Y.C.; Sava, V.M.; Makan, S.Y.; Chen, T.H.J.; Hong, M.Y.; Huang, G.S. Antioxidant activity of melanins derived from tea: Comparison between different oxidative states. Food Chem. 2002, 78, 233–240. [Google Scholar] [CrossRef]
- Barros, L.; Calhelha, R.C.; Vaz, J.A.; Ferreira, I.C.F.R.; Baptista, P.; Estevinho, L.M. Antimicrobial activity and bioactive compounds of Portuguese wild edible mushrooms methanolic extracts. Eur. Food Res. Technol. 2006, 225, 151–156. [Google Scholar] [CrossRef]
- Nasiri, F.; Tarzi, B.G.; Bassiri, A.; Hoseini, S.E. Comparative study on some chemical compounds of button mushrooms (Agaricus Bisporus) cap and stipe during the first to third flushes. Ann. Biol. Res. 2012, 3, 5677–5680. [Google Scholar]
- Fischer, G.; Braun, S.; Thissen, R.; Dott, W. FT-IR spectroscopy as a tool for rapid identification and intra-species characterization of airborne filamentous fungi. J. Microbiol. Methods 2006, 64, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Zervakis, G.I.; Bekiaris, G.; Tarantilis, P.; Pappas, C.S. Rapid strain classification and taxa delimitation within the edible mushroom genus Pleurotus through the use of diffuse reflectance infrared Fourier transform (DRIFT) spectroscopy. Fungal Biol. 2012, 116, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.X.; Liu, G.; Ou, Q.H.; Yu, H.C.; Li, H.M.; Liu, Y. Discrimination of Seven Species of Boletus with Fourier Transform Infrared Spectroscopy. Spectrosc. Spectr. Anal. 2016, 36, 2479–2486. [Google Scholar]
- Mohaček-Grošev, V.; Božac, R.; Puppels, G.J. Vibrational spectroscopic characterization of wild growing mushrooms and toadstools. Spectrochim. Acta Part A 2001, 57, 2815–2829. [Google Scholar] [CrossRef]
- Mellado-Mojica, E.; Lopez, M.G. Identification, classification, and discrimination of agave syrups from natural sweeteners by infrared spectroscopy and HPAEC-PAD. Food Chem. 2015, 167, 349–357. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, A.; Downey, G.; Gowen, A.A.; Barry-Ryan, C.; Frias, J.M. Use of Fourier transform infrared spectroscopy and chemometric data analysis to evaluate damage and age in mushrooms (Agaricus bisporus) grown in Ireland. J. Agric. Food Chem. 2010, 58, 7770–7776. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Li, T.; Liu, H.; Li, J.; Wang, Y. Geographical traceability of wild Boletus edulis based on data fusion of FT-MIR and ICP-AES coupled with data mining methods (SVM). Spectrochim. Acta Part A 2017, 177, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Falandysz, J.; Frankowska, A.; Jarzynska, G.; Dryzalowska, A.; Kojta, A.K.; Zhang, D. Survey on composition and bioconcentration potential of 12 metallic elements in King Bolete (Boletus edulis) mushroom that emerged at 11 spatially distant sites. J. Environ. Sci. Health Part B 2011, 46, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Nnorom, I.C.; Jarzyńska, G.; Drewnowska, M.; Dryżałowska, A.; Kojta, A.; Pankavec, S.; Falandysz, J. Major and trace elements in sclerotium of Pleurotus tuber-regium (Ósū) mushroom-Dietary intake and risk in southeastern Nigeria. J. Food Compos. Anal. 2013, 29, 73–81. [Google Scholar] [CrossRef]
- Medyk, M.; Chudzinska, M.; Baralkiewicz, D.; Falandysz, J. Specific accumulation of cadmium and other trace elements in Sarcodon imbricatus using ICP-MS with a chemometric approach. J. Environ. Sci. Health Part B 2017, 52, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Zhao, Y.; Li, Z.; Li, T.; Wang, Y. Characteristic Fingerprint Based on Low Polar Constituents for Discrimination of Wolfiporia extensa according to Geographical Origin Using UV Spectroscopy and Chemometrics Methods. J. Anal. Methods Chem. 2014, 2014, 519424. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xie, M.Y.; Yan, Y.; Zhu, S.B.; Nie, S.P.; Li, C.; Wang, Y.X.; Gong, X.F. Discrimination of Ganoderma lucidum according to geographical origin with near infrared diffuse reflectance spectroscopy and pattern recognition techniques. Anal. Chim. Acta 2008, 618, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Casale, M.; Bagnasco, L.; Zotti, M.; Di Piazza, S.; Sitta, N.; Oliveri, P. A NIR spectroscopy-based efficient approach to detect fraudulent additions within mixtures of dried porcini mushrooms. Talanta 2016, 160, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Jin, H.; Liu, H.; Wang, Y. Ultraviolet spectroscopy combined with ultra-fast liquid chromatography and multivariate statistical analysis for quality assessment of wild Wolfiporia extensa from different geographical origins. Spectrochim. Acta Part A 2016, 165, 61–68. [Google Scholar] [CrossRef] [PubMed]


| Geographical Origins | Quantity | Longitude | Latitude | Altitude (m) |
|---|---|---|---|---|
| Potatso national park, Xianggelila, Diqing | 10 | 99.908 | 27.802 | 3515 |
| Midu, Dali | 10 | 100.491 | 25.344 | 1670 |
| Baohe, Weixi, Diqing | 7 | 99.286 | 27.177 | 2300 |
| Longyang, Baoshan | 10 | 99.166 | 25.121 | 1680 |
| Wenshui, Bajie, Anning, Kunming | 9 | 102.393 | 24.577 | 1846 |
| Fengyi, Bajie, Anning, Kunming | 10 | 102.333 | 24.692 | 1984 |
| Tongchang, Yimen, Yuxi | 7 | 102.039 | 24.714 | 2198 |
| Songgui, Heqing, Dali | 6 | 100.210 | 26.354 | 1944 |
| Dongshan, Wenshan | 6 | 104.281 | 23.400 | 1430 |
| Liujie, Bajie, Anning, Kunming | 8 | 102.686 | 24.532 | 1998 |
| Suoyishan, Weize, Shilin, Kunming | 9 | 103.346 | 24.645 | 1893 |
| Model | RMSEE | RMSECV | Accuracy of Calibration Set | Accuracy of Validation Set |
|---|---|---|---|---|
| Cap | 0.076 | 0.251 | 100.000% | 90.625% |
| Stipe | 0.079 | 0.244 | 100.000% | 96.875% |
| Model | C | γ | Accuracy of Calibration Set | Accuracy of Test Set |
|---|---|---|---|---|
| Cap | 1.000 | 0.044194 | 100.000% | 100.000% |
| Stipe | 1.000 | 0.0625 | 100.000% | 100.000% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, L.; Liu, H.; Li, J.; Li, T.; Wang, Y. Feature Fusion of ICP-AES, UV-Vis and FT-MIR for Origin Traceability of Boletus edulis Mushrooms in Combination with Chemometrics. Sensors 2018, 18, 241. https://doi.org/10.3390/s18010241
Qi L, Liu H, Li J, Li T, Wang Y. Feature Fusion of ICP-AES, UV-Vis and FT-MIR for Origin Traceability of Boletus edulis Mushrooms in Combination with Chemometrics. Sensors. 2018; 18(1):241. https://doi.org/10.3390/s18010241
Chicago/Turabian StyleQi, Luming, Honggao Liu, Jieqing Li, Tao Li, and Yuanzhong Wang. 2018. "Feature Fusion of ICP-AES, UV-Vis and FT-MIR for Origin Traceability of Boletus edulis Mushrooms in Combination with Chemometrics" Sensors 18, no. 1: 241. https://doi.org/10.3390/s18010241





