Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels
Abstract
:1. Introduction
2. Circulating Breast Cancer Biomarkers
3. Electrochemical Biosensing of Circulating Breast Cancer Biomarkers
3.1. Electrochemical Biosensing of Gene Specific Mutations and miRNAs Associated with Breast Cancer in Biofluids
3.2. Electrochemical Biosensing of Breast Cancer Protein Circulating Biomarkers
3.2.1. Electrochemical Aptasensors for Breast Cancer Protein Circulating Biomarkers
3.2.2. Electrochemical Immunosensors for Protein Circulating Biomarkers
3.2.3. Electrochemical Peptide Biosensor for Breast Cancer Protein Circulating Biomarkers
3.3. Electrochemical Biosensing of Circulating Breast Cancer Cells
3.4. Electrochemical Biosensing for Multiple Determination of Circulating Breast Cancer Biomarkers
4. General Considerations, Challenges and Future Prospects
Acknowledgments
Conflicts of Interest
References
- Mansor, N.A.; Zain, Z.M.; Hamzah, H.H.; Noorden, M.S.A.; Jaapar, S.S.; Beni, V.; Ibupoto, Z.H. Detection of breast cancer 1 (BRCA1) gene using an electrochemical DNA biosensor based on immobilized ZnO nanowires. Open J. Appl. Biosens. 2014, 3, 9–17. [Google Scholar] [CrossRef]
- Wang, W.; Fan, X.; Xu, S.; Davis, J.J.; Luo, X. Low fouling label-free DNA sensor based on polyethylene glycols decorated with gold nanoparticles for the detection of breast cancer biomarkers. Biosens. Bioelectron. 2015, 71, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.R.; Moreira, F.T.C.; Fernandes, R.; Sales, M.G. Novel and simple electrochemical biosensor monitoring attomolar levels of miRNA-155 in breast cancer. Biosens. Bioelectron. 2016, 80, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Moscovici, M.; Bhimji, A.; Kelley, S.O. Rapid and specific electrochemical detection of prostate cancer cells using an aperture sensor array. Lab Chip 2013, 13, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chandra, P.; Shim, Y.-B. Ultrasensitive and selective electrochemical diagnosis of breast cancer based on a hydrazine-Au nanoparticle-aptamer bioconjugate. Anal. Chem. 2013, 85, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Wang, B.; Yu, Z.; Zhao, J.; Tang, Y. Ultrasensitive electrochemical detection of microRNA with star trigon structure and endonuclease mediated signal amplification. Biosens. Bioelectron. 2015, 63, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, M.; Rahaiea, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar] [CrossRef] [PubMed]
- NCI Dictionary of Cancer Terms. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms?cdrid=45618 (accessed on 6 July 2017).
- Costa Rama, E.; Costa-García, A. Screen-printed electrochemical immunosensors for the detection of cancer and cardiovascular biomarkers. Electroanalysis 2016, 28, 1700–1715. [Google Scholar] [CrossRef]
- Rawiwan, L. Development of electrochemical immunosensors towards point-of-care cancer diagnostics: Clinically relevant studies. Electroanalysis 2016, 28, 1716–1729. [Google Scholar] [CrossRef]
- Sezginturk, M.K. New impedimetric biosensor utilizing VEGF receptor-1 (Flt-1): Early diagnosis of vascular endothelial growth factor in breast cancer. Biosens. Bioelectron. 2011, 26, 4032–4039. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Electrochemical genosensing of circulating biomarkers. Sensors 2017, 17, 866. [Google Scholar] [CrossRef] [PubMed]
- Low, K.-F.; Rijiravanich, P.; Singh, K.K.B.; Surareungchai, W.; Yean, C.Y. An electrochemical genosensing assay based on magnetic beads and gold nanoparticle-loaded latex microspheres for Vibrio cholera detection. J. Biomed. Nanotechnol. 2015, 11, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Ilkhani, H.; Sarparast, M.; Noori, A.; Bathaie, S.Z.; Mousavi, M.F. Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker, using gold nanoparticles as a signaling probe. Biosens. Bioelectron. 2015, 74, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.M.; Gonçalves, I.C.; Magalhães, F.D. Graphene-based materials biocompatibility: A review. Colloids Surf. B Biointerface 2013, 111, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, N.J.; Okon, S.L. Nanomaterial-based electrochemical immunosensors for clinically significant biomarkers. Materials 2014, 7, 4669–4709. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, P.A.; Sandhyarani, N. Graphene-DNA electrochemical sensor for the sensitive detection of BRCA1 gene. Sens. Actuator B Chem. 2014, 204, 777–782. [Google Scholar] [CrossRef]
- Benvidi, A.; Tezerjani, M.D.; Jahanbani, S.; Ardakani, M.M.; Moshtaghioun, S.M. Comparison of impedimetric detection of DNA hybridization on the various biosensors based on modified glassy carbon electrodes with PANHS and nanomaterials of RGO and MWCNTs. Talanta 2016, 147, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Dehghani Firouzabadi, A.; Dehghan Tezerjani, M.; Moshtaghiun, S.M.; Mazloum-Ardakani, M.; Ansarin, A. A highly sensitive and selective electrochemical DNA biosensor to diagnose breast cancer. J. Electroanal. Chem. 2015, 750, 57–64. [Google Scholar] [CrossRef]
- Shuai, H.-L.; Huang, K.-J.; Xing, L.-L.; Chen, Y.-X. Ultrasensitive electrochemical sensing platform for microRNA based on tungsten oxide-graphene composites coupling with catalyzed hairpin assembly target recycling and enzyme signal amplification. Biosens. Bioelectron. 2016, 86, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Wang, W.; Kang, T.; Liu, J.; Shiu, K.-K.; Leung, C.-H.; Ma, D.-L. Ultrasensitive electrochemical detection of miRNA-21 by using an Iridium(III) complex as catalyst. Biosens. Bioelectron. 2016, 86, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Khan, N.; Ghobadloo, S.M.; Cheng, J.; Pezacki, J.P.; Berezovski, M.V. Three-mode electrochemical sensing of ultralow microRNA levels. J. Am. Chem. Soc. 2013, 135, 3027–3038. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-Y.; Chen, X.; Liu, T.; Li, J.; Yang, H.-H.; Chen, J.-H.; Chen, G.-N. Ultrasensitive electrochemical detection of cancer-associated circulating microRNA in serum samples based on DNA concatamers. Biosens. Bioelectron. 2013, 50, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Z.; Cai, S.; Wen, F.; Wu, D.; Liu, Y.; Wu, F.; Lan, J.; Han, Z.; Chen, J. An electrochemical microRNA biosensor based on protein p19 combining an acridone derivate as indicator and DNA concatamers for signal amplification. Electrochem. Commun. 2015, 60, 185–189. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, Y.; Wei, X.; Huang, Y.; Liu, L. An electrochemical microRNAs biosensor with the signal amplification of alkaline phosphatase and electrochemical–chemical–chemical redox cycling. Anal. Chim. Acta 2015, 878, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.-F.; Jiang, N.; Li, X.; Zhang, L.; Hu, L.; Chen, X.; Jiang, L.-P.; Abdel-Halim, E.S.; Zhu, J.-J. Target-triggered triple isothermal cascade amplification strategy for ultrasensitive microRNA-21 detection at sub-attomole level. Biosens. Bioelectron. 2016, 85, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, D.-Z.; Cai, S.-X.; Chen, M.; Xia, Y.-K.; Wu, F.; Chen, J.-H. An immobilization-free electrochemical impedance biosensor based on duplex-specific nuclease assisted target recycling for amplified detection of microRNA. Biosens. Bioelectron. 2016, 75, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Shen, B.; Yuan, R.; Cheng, W.; Xu, H.; Ding, S. An electrochemical biosensor for highly sensitive determination of microRNA based on enzymatic and molecular beacon mediated strand displacement amplification. J. Electroanal. Chem. 2015, 756, 147–152. [Google Scholar] [CrossRef]
- Zhao, S.; Yang, W.; Lai, R.Y. A folding-based electrochemical aptasensor for detection of vascular endothelial growth factor in human whole blood. Biosens. Bioelectron. 2011, 26, 2442–2447. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wen, W.; Wang, Q.; Xiong, H.; Zhang, X.; Gu, H.; Wang, S. Novel electrochemical aptamer biosensor based on an enzyme-gold nanoparticle dual label for the ultrasensitive detection of epithelial tumour marker MUC1. Biosens. Bioelectron. 2014, 53, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Salimian, R.; Kékedy-Nagy, L.; Ferapontova, E.E. Specific picomolar detection of a breast cancer biomarker HER-2/neu protein in serum: Electrocatalytically amplified electroanalysis by the aptamer/PEG-modified electrode. ChemElectroChem 2017, 4, 872–879. [Google Scholar] [CrossRef]
- Li, W.; Yuan, R.; Chai, Y.; Chen, S. Reagentless amperometric cancer antigen 15-3 immunosensor based on enzyme-mediated direct electrochemistry. Biosens. Bioelectron. 2010, 25, 2548–2552. [Google Scholar] [CrossRef] [PubMed]
- Munge, B.S.; Coffey, A.L.; Doucette, J.M.; Somba, B.K.; Malhotra, R.; Patel, V.; Gutkind, J.S.; Rusling, J.F. Nanostructured immunosensor for attomolar detection of cancer biomarker Interleukin-8 using massively labeled superparamagnetic particles. Angew. Chem. Int. Ed. 2011, 50, 7915–7918. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tang, D.; Zhang, B.; Liu, B.; Cui, Y.; Chen, G. Electrochemical immunosensor for carcinoembryonic antigen based on nanosilver-coated magnetic beads and gold-graphene nanolabels. Talanta 2012, 91, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Elshafey, R.; Tavares, A.C.; Siaj, M.; Zourob, M. Electrochemical impedance immunosensor based on gold nanoparticles-protein for the detection of cancer marker epidermal growth factor receptor in human plasma and brain tissue. Biosens. Bioelectron. 2013, 50, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Sonuç, M.N.; Sezgintürk, M.K. Ultrasensitive electrochemical detection of cancer associated biomarker HER3 based on anti-HER3 biosensor. Talanta 2014, 120, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Marques, R.C.B.; Viswanathan, S.; Nouws, H.P.A.; Delerue-Matos, C.; González-García, M.B. Electrochemical immunosensor for the analysis of the breast cancer biomarker HER2 ECD. Talanta 2014, 129, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Emami, M.; Shamsipur, M.; Saber, R.; Irajirad, R. An electrochemical immunosensor for detection of a breast cancer biomarker based on antiHER2–iron oxide nanoparticle bioconjugates. Analyst 2014, 139, 2858–2866. [Google Scholar] [CrossRef] [PubMed]
- Samanman, S.; Numnuam, A.; Limbut, W.; Kanatharana, P.; Thavarungkul, P. Highly-sensitive label-free electrochemical carcinoembryonic antigen immunosensor based on a novel Au nanoparticles-graphene-chitosan nanocomposite cryogel electrode. Anal. Chim. Acta 2015, 853, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.-S.; Li, X.-Y.; Xie, Z.-H.; Li, X.-G.; Zhang, H.-Y. Poly(o-phenylenediamine) nanosphere-conjugated capture antibody immobilized on a glassy carbon electrode for electrochemical immunoassay of carcinoembryonic antigen. Microchim. Acta 2015, 182, 2541–2549. [Google Scholar] [CrossRef]
- Han, J.; Jiang, L.; Li, F.; Wang, P.; Liu, Q.; Dong, Y.; Li, Y.; Wei, Q. Ultrasensitive non-enzymatic immunosensor for carcino-embryonic antigen based on palladium hybrid vanadium pentoxide/multiwalled carbon nanotubes. Biosens. Bioelectron. 2016, 77, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Eletxigerra, U.; Martinez-Perdiguero, J.; Merino, S.; Barderas, R.; Torrente-Rodríguez, R.M.; Villalonga, R.; Pingarrón, J.M.; Campuzano, S. Amperometric magnetoimmunosensor for ErbB2 breast cancer biomarker determination in human serum, cell lysates and intact breast cancer cells. Biosens. Bioelectron. 2015, 70, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Eletxigerra, U.; Martinez-Perdiguero, J.; Merino, S.; Barderas, R.; Ruiz-Valdepeñas Montiel, V.; Villalonga, R.; Pingarrón, J.M.; Campuzano, S. Estrogen receptor α determination in serum, cell lysates and breast cancer cells using an amperometric magnetoimmunosensing platform. Sens. Bio-Sens. Res. 2016, 7, 71–76. [Google Scholar] [CrossRef]
- Peng, D.; Liang, R.-P.; Huang, H.; Qiu, J.-D. Electrochemical immunosensor for carcinoembryonic antigen based on signal amplification strategy of graphene and Fe3O4/Au NPs. J. Electroanal. Chem. 2016, 761, 112–117. [Google Scholar] [CrossRef]
- Zheng, L.; Niu, X.; Zhao, J.; Liu, T.; Liu, Y.; Yang, Y. A Label-Free Immunosensor for CEA based on Pd–Ir bimetallic nanoparticles. J. Nanosci. Nanotechnol. 2016, 16, 5984–5990. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Gao, Y.; Liu, Z.; Xu, J.; Lu, L.; Yu, Y. Three-dimensional gold nanoparticles/Prussian Blue-poly(3,4-ethylenedioxythiophene) nanocomposite as novel redox matrix for label-free electrochemical immunoassay of carcinoembryonic antigen. Sens. Actuator B Chem. 2017, 239, 76–84. [Google Scholar] [CrossRef]
- Wang, K.; He, M.-Q.; Zhai, F.-H.; He, R.-H.; Yu, Y.-L. A novel electrochemical biosensor based on polyadenine modified aptamer for label-free and ultrasensitive detection of human breast cancer cells. Talanta 2017, 166, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhu, L.; Guo, C.; Gao, T.; Zhu, X.; Li, G. A new electrochemical method for the detection of cancer cells based on small molecule-linked DNA. Biosens. Bioelectron. 2013, 49, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Song, P.; Zhou, G.; Zuo, X.; Aldalbahi, A.; Lou, X.; Shi, J.; Fan, C. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 2016, 11, 1244–1263. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, L.; Li, G.; Jing, F.; Mao, H.; Jin, Q.; Zhai, W.; Zhang, H.; Zhao, J.; Jia, C. Multiplexed detection of lung cancer biomarkers based on quantum dots and microbeads. Talanta 2016, 156, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jiang, W.; Dai, S.; Wang, L. Multiplexed detection of cytokines based on dual bar-code strategy and single-molecule counting. Anal. Chem. 2016, 88, 1578–1584. [Google Scholar] [CrossRef] [PubMed]
- Munge, B.S.; Stracensky, T.; Gamez, K.; DiBiase, D.; Rusling, J.F. Multiplex immunosensor arrays for electrochemical detection of cancer biomarker proteins. Electroanalysis 2016, 28, 2644–2658. [Google Scholar] [CrossRef] [PubMed]
- Topkaya, S.N.; Azimzadeh, M.; Ozsoz, M. Electrochemical biosensors for cancer biomarkers detection: Recent advances and challenges. Electroanalysis 2016, 28, 1402–1419. [Google Scholar] [CrossRef]
- Wei, F.; Patel, P.; Liao, W.; Chaudhry, K.; Zhang, L.; Arellano-Garcia, M.; Hu, S.; Elashoff, D.; Zhou, H.; Shukla, S.; et al. Electrochemical sensor for multiplex biomarkers detection. Clin. Cancer Res. 2009, 15, 4446–4452. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Campuzano, S.; Ruiz-Valdepeñas Montiel, V.; Gamella, M.; Pingarrón, J.M. Electrochemical bioplatforms for the simultaneous determination of interleukin (IL)-8 mRNA and IL-8 protein oral cancer biomarkers in raw saliva. Biosens. Bioelectron. 2016, 77, 543–548. [Google Scholar] [CrossRef]
- Eletxigerra, U.; Martinez-Perdiguero, J.; Merino, S.; Barderas, R.; Ruiz-Valdepeñas Montiel, V.; Villalonga, R.; Pingarrón, J.M.; Campuzano, S. Electrochemical magnetoimmunosensor for progesterone receptor determination. Application to the simultaneous detection of estrogen and progesterone breast-cancer related receptors in raw cell lysates. Electroanalysis 2016, 28, 1787–1794. [Google Scholar] [CrossRef]
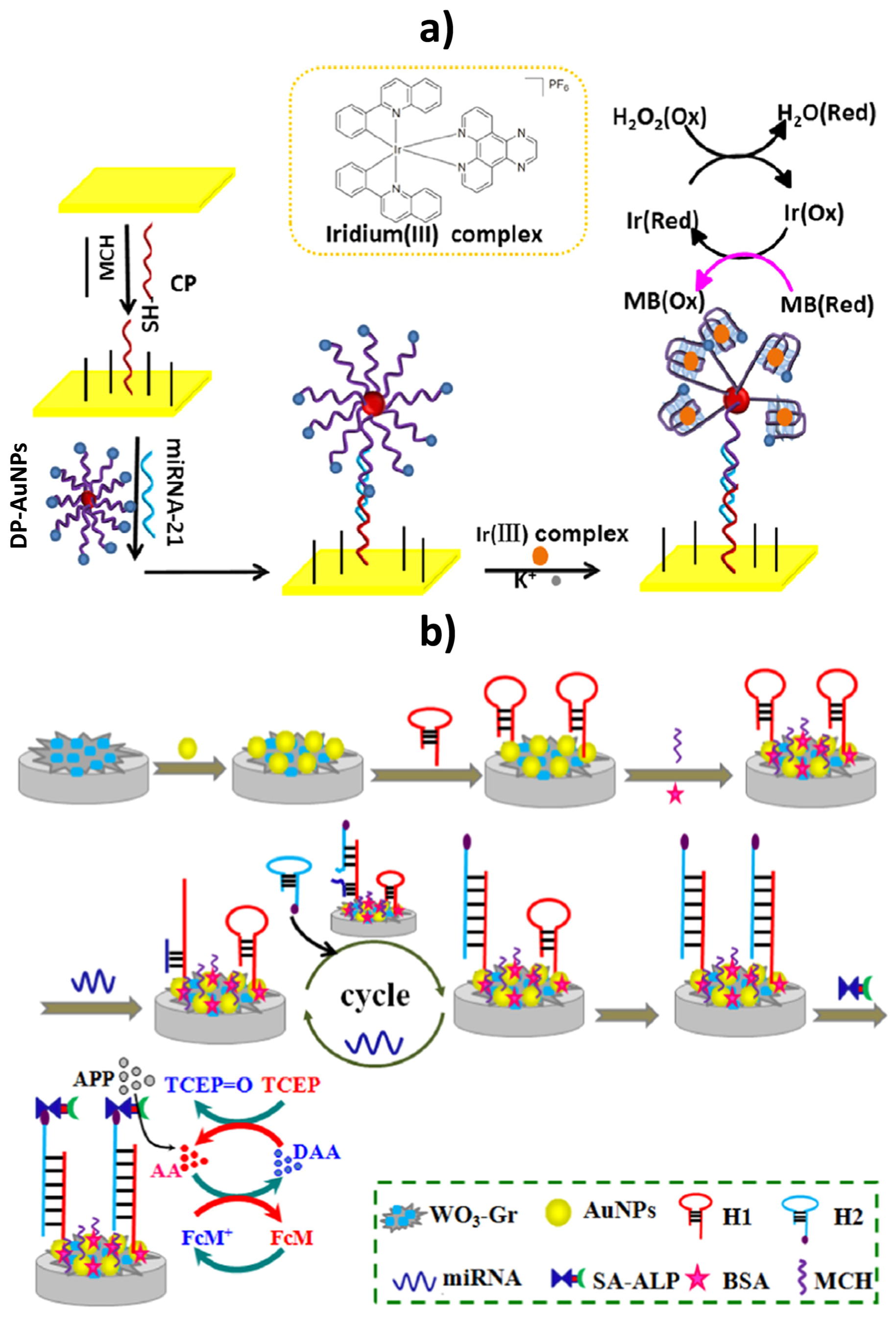
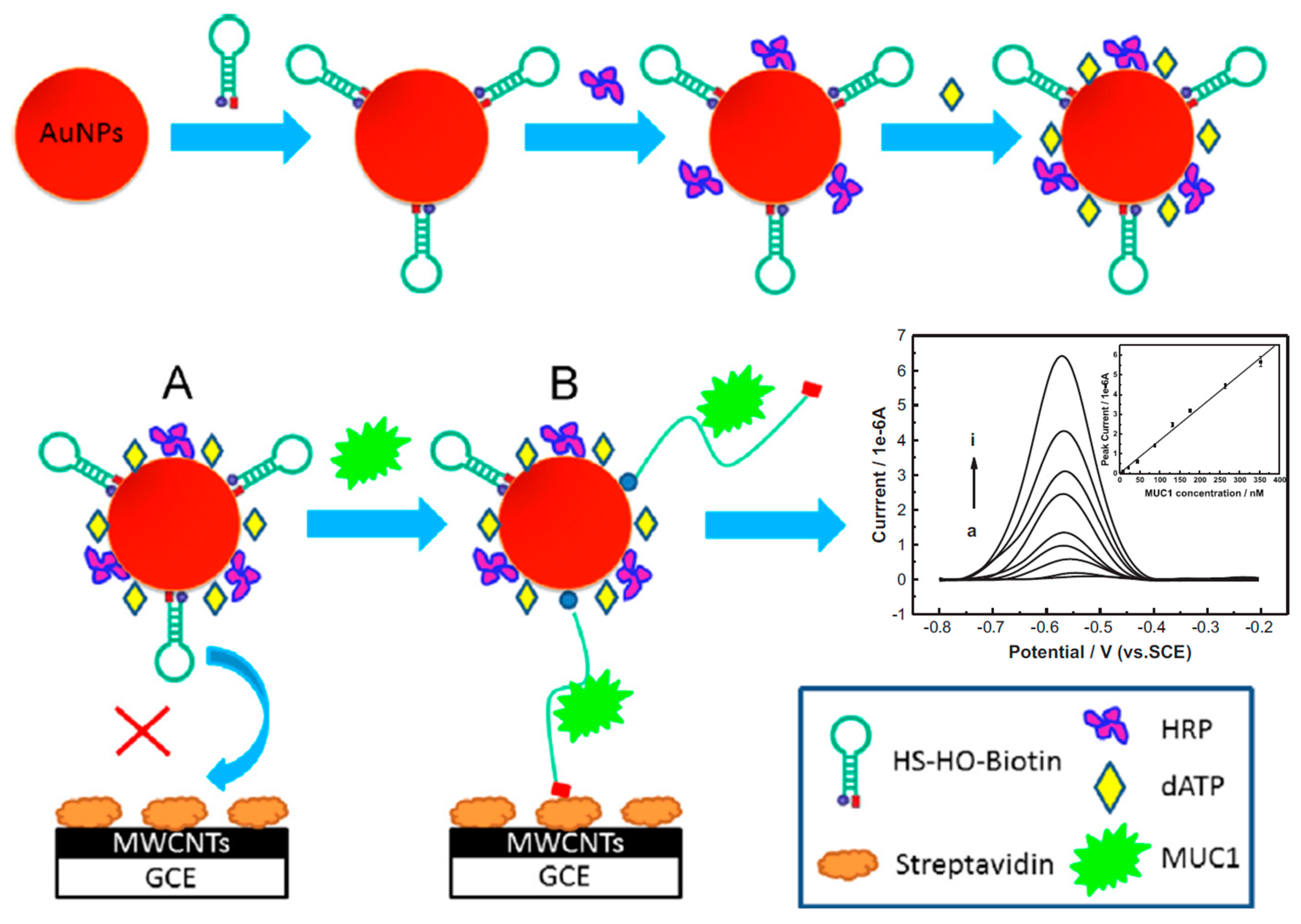

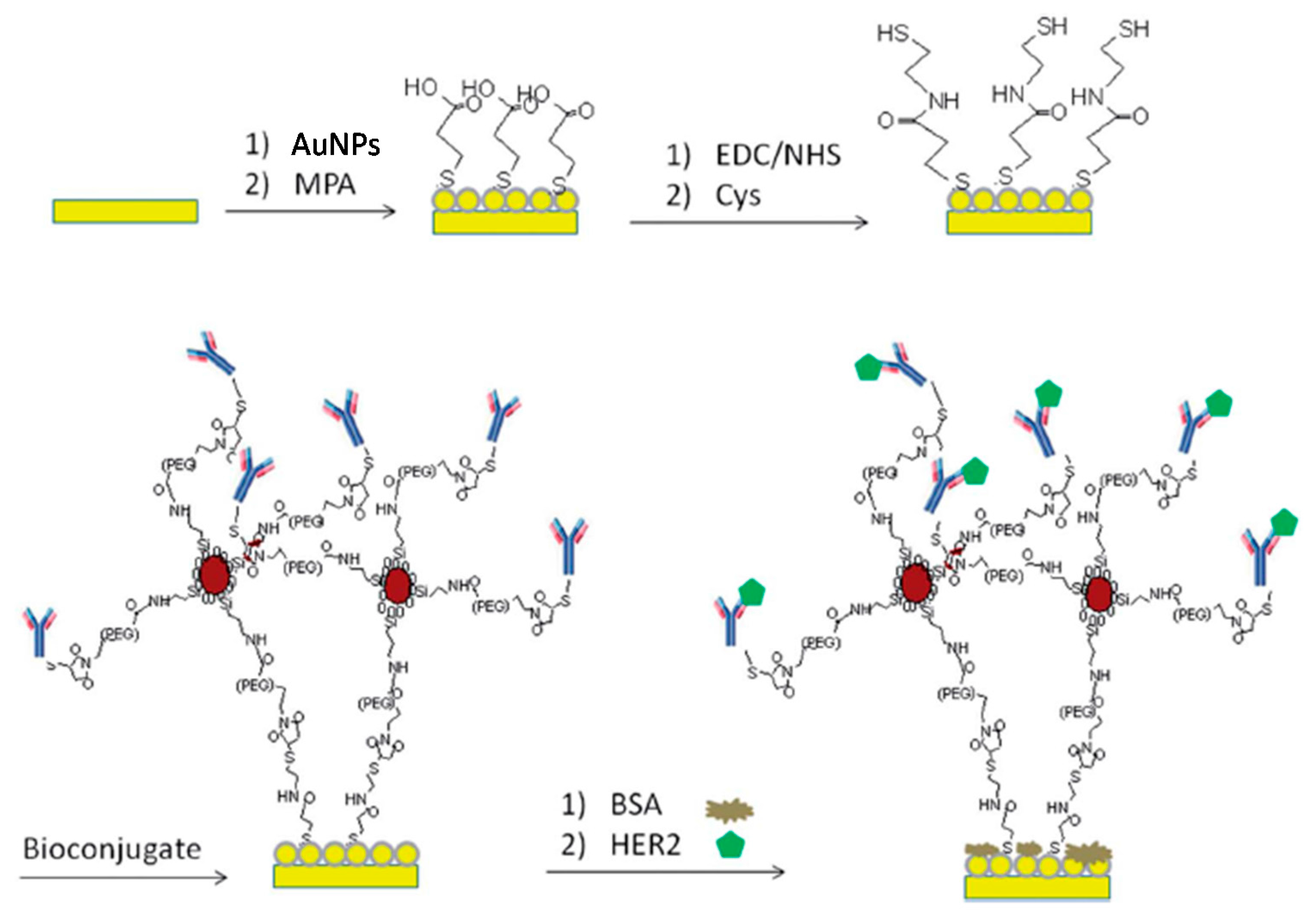

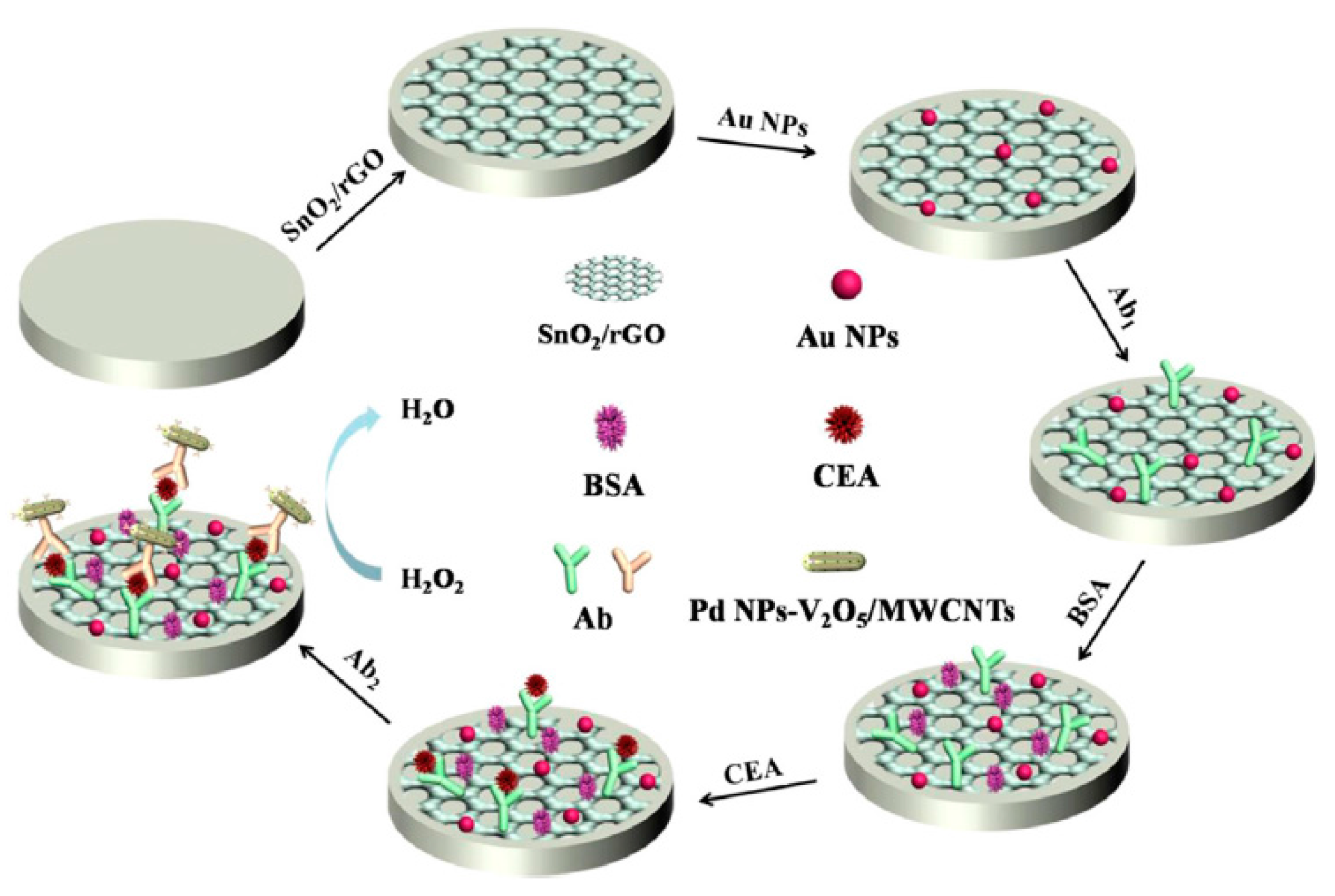
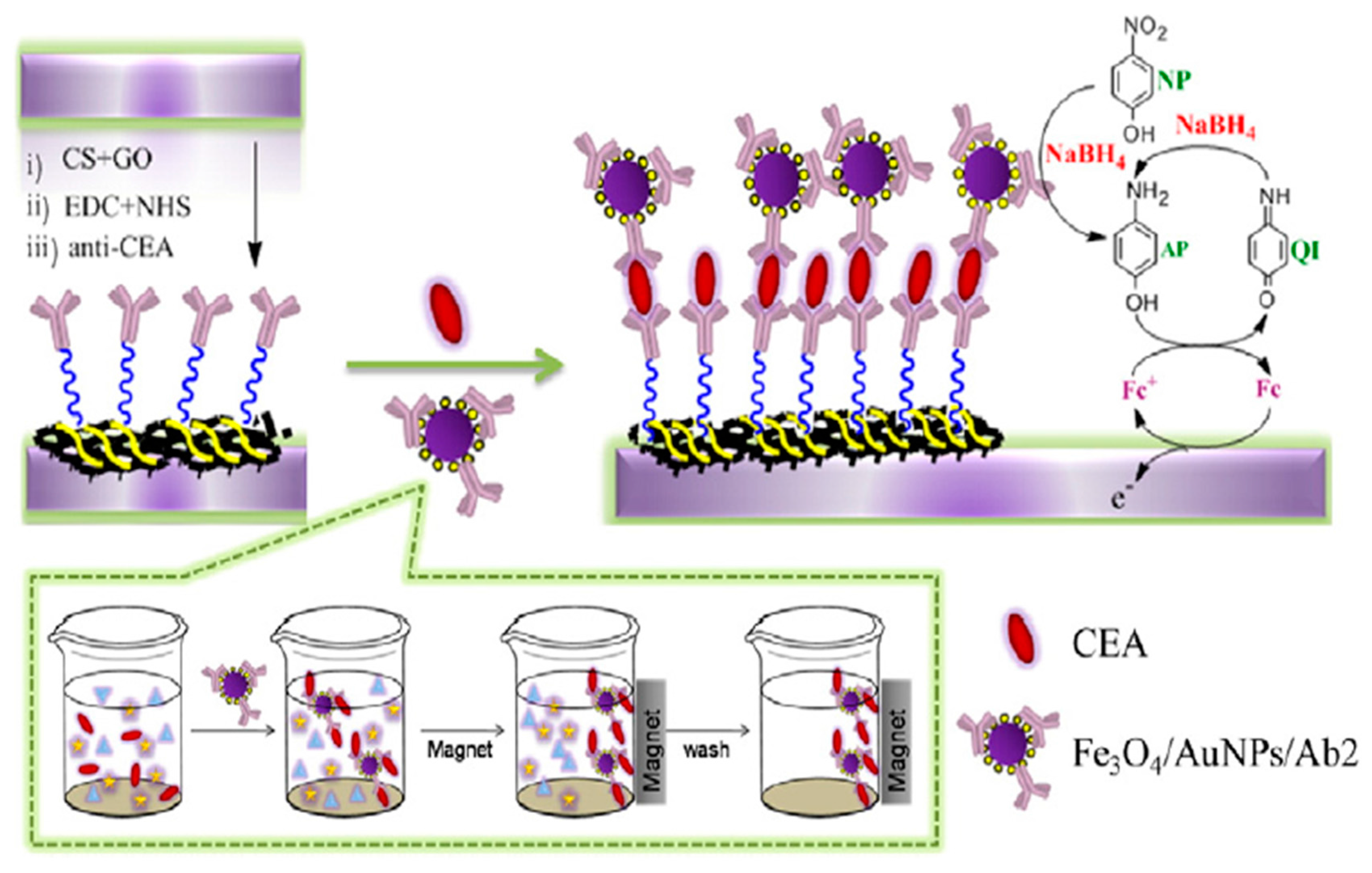
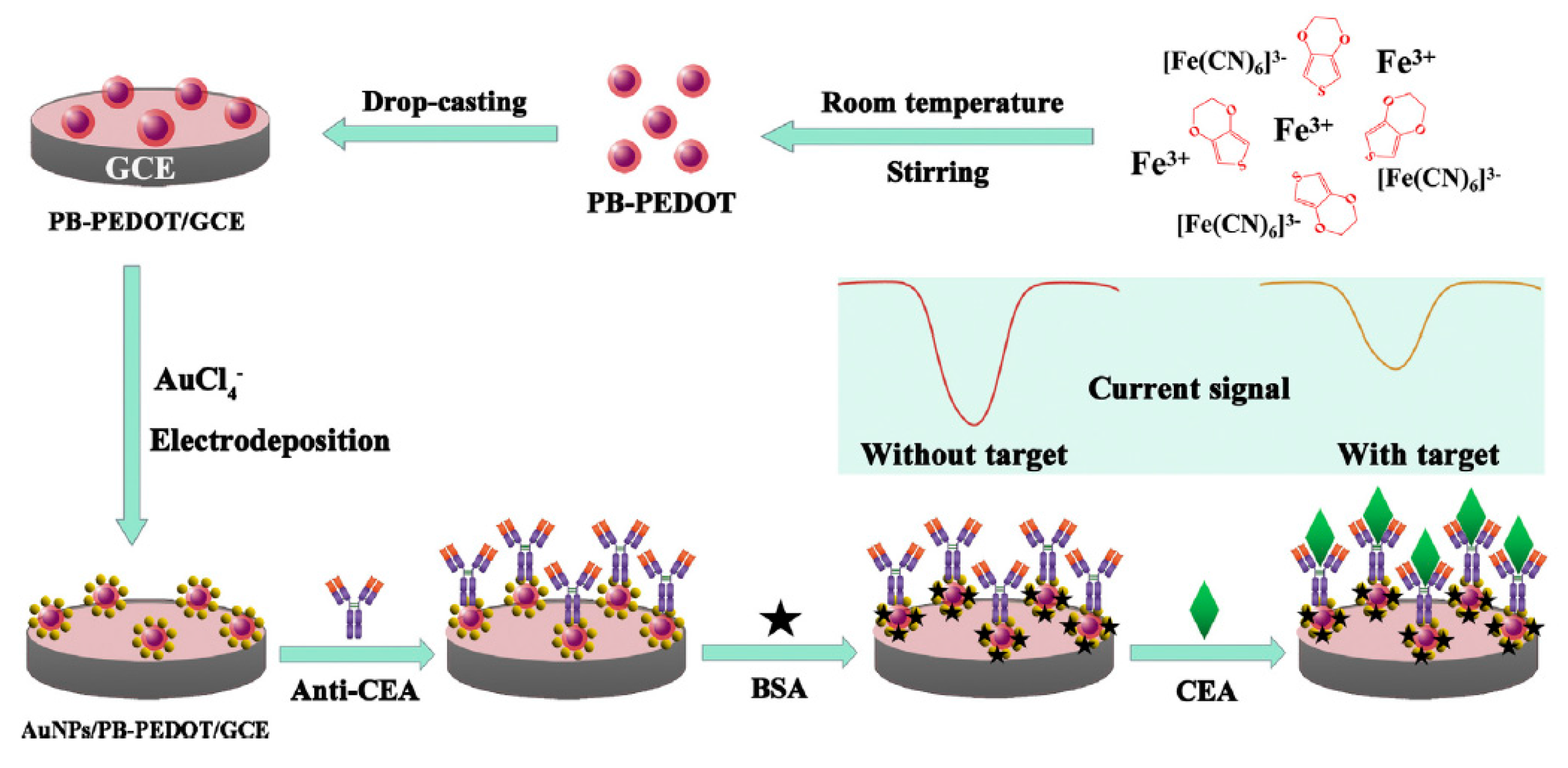

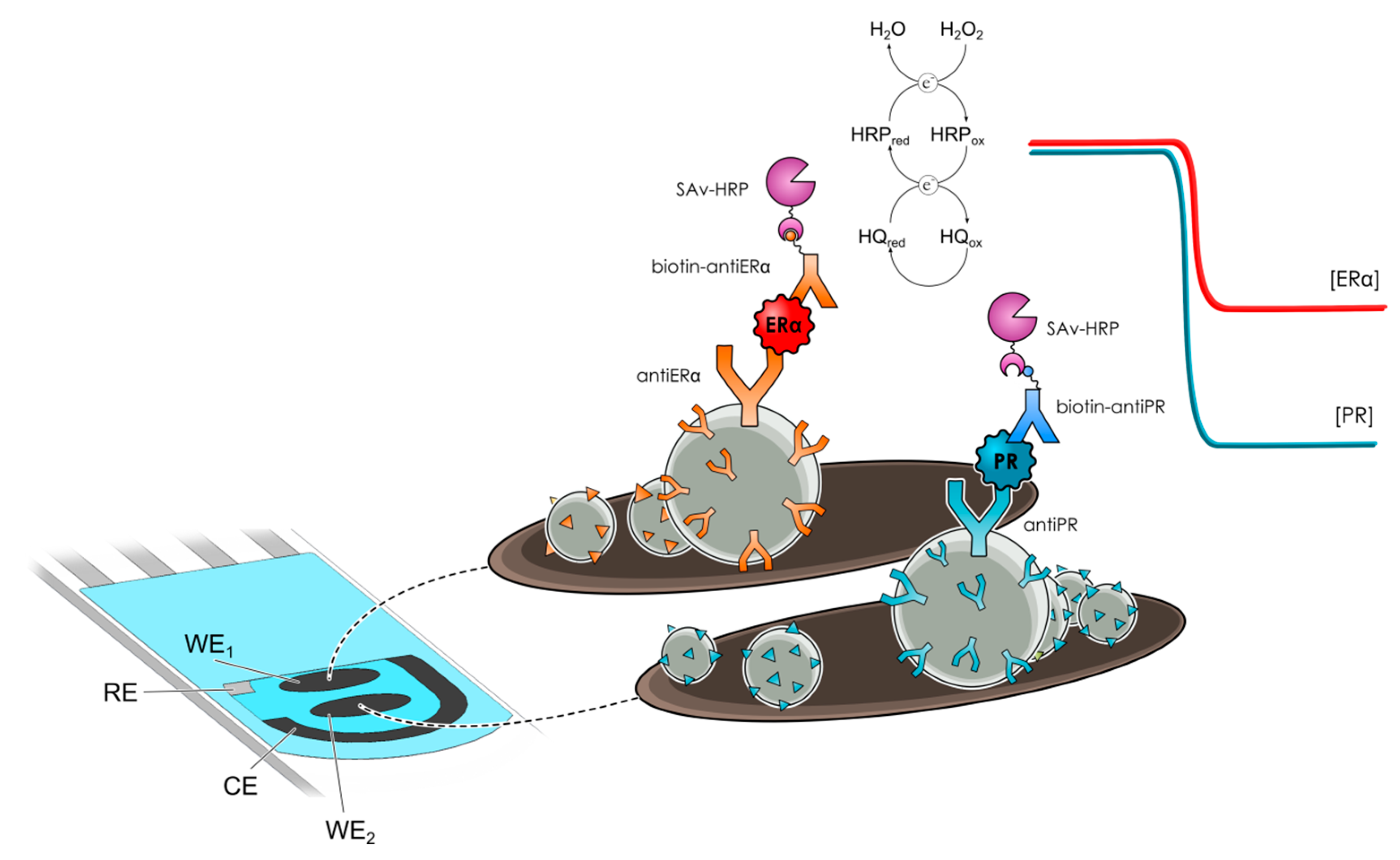
| Electrode | Method | Target Oligonucleotide | Electrochemical Technique/Redox Probe | L.R. | LOD | Applicability | Ref. |
|---|---|---|---|---|---|---|---|
| Gold electrode | Immobilization of a binary self-assembled monolayer composed of a thiolated specific capture DNA probe (BRCA1 5382 insC mutation detection) and 6-mercapto-1-hexanol (MCH) on the gold electrode. | BRCA1 gene mutations | EIS/[Fe(CN)6]4−/3− | 1.0 × 10−19‒1.0 × 10−7 M | 4.6 × 10−20 M | Genomic DNA extracted from peripheral blood samples | [19] |
| GCE | Preparation of an antifouling GCE by highly cross-linking of polyethylene glycol films containing amine groups where AuNPs were self-assembled for further immobilization of a BRCA1 related 19-mer DNA sequence. | BRCA1 gene mutations | EIS/[Fe(CN)6]4−/3− | 50.0 fM‒1.0 nM | 1.72 fM | Doped serum samples | [2] |
| AuNPs-modified SPCE | Immobilization of a thiolated RNA probe, p19 binding onto the RNA-RNA duplex formed on the electrode surface by direct hybridization, and displacemen t of the attached p19 by incubation in a mixture of a target miRNA and a nonthiolated RNA probe at high concentration. | miRNA-21, miRNA-32, miRNA-122 | SWV/K3[Fe(CN)6] and [Ru(NH3)6]Cl3 | 10 aM–1 μM | 5 aM | Human serum samples | [22] |
| Au-SPE | Immobilization of a thiolated RNA probe and direct hybridization. | miRNA-155 | SWV/[Fe(CN)6]3−/4− | 10 aM–1.0 nM | 5.7 aM | Human serum samples | [3] |
| GCE functionalized with AuNRs decorated on GO sheets | Immobilization of a thiolated RNA probe and direct hybridization. | miRNA-155 | DPV/OB | 2.0 fM–8.0 pM | 0.6 fM | Spiked human plasma samples | [7] |
| Au-SPE | Immobilization of a thiolated capture probe on the gold electrode. In the presence of the target miRNA the stem-loop structure of such capture probe is unfolded and hybridizes with the DNA concatamers. | miRNA-21 | DPV/[Ru(NH3)6]3+ | 100 aM–100 pM | 100 aM | Human serum samples | [23] |
| GCE | Bi-functional Janus probe containing both the complementary RNA oligonucleotide sequence for the target miRNA and the DNA sequence used as primer for long-range self-assembled DNA concatamers. The RNA duplexes generated by hybridization between the Janus probe and the target miRNA was selectively captured onto the surface of p19-MBs and long DNA concatamers anchored to the MBs were regenerated by addition of two specific auxiliary probes. | miRNA-21 | SWV/DSA intercalated | 20 aM–100 aM | 6 aM | Human serum samples | [24] |
| GCE modified with AuNPs using PDDA | Immobilization of a thiolated capture probe with MB labeled at 5′ end. Hybridization with the target miRNA in the presence of an auxiliary probe, forming a star trigon structure on the electrode surface. The endonuclease cleaves the capture probe on capture/auxiliary probes duplex, releasing microRNA and auxiliary probe back to the solution. | miRNA-21 | SWV/MB of the AP | 100 aM–1 nM | 30 aM | Human serum samples | [6] |
| Gold electrode | Immobilization of a hairpin-like DNA probe modified with a thiol and a biotin on a gold electrode through the thiolated moiety. After hybridization with the target miRNA the biotin group in the capture probe was forced away from the electrode surface, allowing for the coupling of Strept-ALP. | miRNA-21 | Amperometry/AA + FcM + TCEP | 0.5 fM–1 pM | 0.2 fM | Human serum samples | [25] |
| Gold electrode | Immobilization of a thiolated capture probe on the gold electrode. In the presence of miRNA-21, a sandwiched DNA complex was formed between the capture and a MB-labeled G-rich detection probe attached onto AuNPs. Upon addition of K+, the structure of the detector probe changed to a G-quadruplex and the iridium(III) complex could selectively interact with it and catalyze the reduction of H2O2, in the presence of MB. | miRNA-21 | CV/H2O2 + MB | 5.0 fM–1.0 pM | 1.6 fM | Spiked human serum samples | [21] |
| Tungsten oxide (WO3)-graphene composites coupled with AuNPs | Immobilization of a thiol-terminated capture probe H1 immobilized on the electrode through Au–S interaction. Hybridization with the target miRNA opens H1 hairpin structure and another stable biotinylated hairpin DNA (H2) displaced target miRNA releasing it back to the sample solution for the next cycle. After this cyclic process a large amount of H1-H2 duplex was produced and a lot of Strept-ALP molecules were immobilized on the electrode. | miRNA-21 | DPV/AA + FcM + TCEP | 0.1 fM–100 pM | 0.05 fM | Human serum samples | [20] |
| GCE | Target recycling, nicking-replication reaction and DNAzyme catalysis coupling. | miRNA-21 | Amperometry/TMB+ H2O2 | 1 aM–100 pM | 0.5 aM | Spiked human serum samples | [26] |
| MGCE | Method based on both the DSNATR and capture probes enriched from the solution to the electrode surface using MBs. In the absence of the target miRNA, the capture probes cannot be hydrolyzed due to the low activity of duplex-specific nuclease against ss-DNA, the intact capture probes could be attached to Strep-MBs and hence onto the surface of a magnetic-GCE, resulting in a compact negatively charged layer, giving rise to a large charge-transfer resistance measured in the presence of [Fe(CN)6]4−/3−. Conversely, in the presence of miRNA-21, it hybridized with the capture probes to form a DNA-RNA heteroduplex and the DSN hydrolyzed the target-binding part of the capture probe thus liberating the intact miRNA-21, which was able to trigger the permanent hydrolysis of multiple capture probes which were, finally, all digested. Therefore, the negatively charged layer could not be formed and a small charge-transfer resistance was measured. | miRNA-21 | EIS/[Fe(CN)6]4−/3− | 0.5–40 fM | 60 aM | Human serum samples | [27] |
| Gold electrode | The molecular beacon template consisted of three domains: a miRNA-binding domain, a recognition domain by Nb.BbvCI, and an amplification domain for producing DNA triggers. In the presence of target miRNA, the specific hybridization with the corresponding domain opened the hairpin structure of the molecular beacon template, which led to a part duplex. Then, the target miRNA was extended along the template to form a complete duplex by Klenow fragment and dNTPs. Subsequently, the nicking enzyme specifically recognized the duplex nicking site, cleaving the upper extended DNA strand and exposing a new replication site for polymerase. While one part of DNA triggers bound to the capture probes immobilized on the gold electrode the other part hybridized with the biotinylated detector probe, which could be linked to Strep-ALP. | miRNA-222 | DPV/α-NP | 50 pM–10 nM | 40 pM | Spiked serum samples | [28] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels. Sensors 2017, 17, 1993. https://doi.org/10.3390/s17091993
Campuzano S, Pedrero M, Pingarrón JM. Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels. Sensors. 2017; 17(9):1993. https://doi.org/10.3390/s17091993
Chicago/Turabian StyleCampuzano, Susana, María Pedrero, and José Manuel Pingarrón. 2017. "Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels" Sensors 17, no. 9: 1993. https://doi.org/10.3390/s17091993
APA StyleCampuzano, S., Pedrero, M., & Pingarrón, J. M. (2017). Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels. Sensors, 17(9), 1993. https://doi.org/10.3390/s17091993







