Linear vs. Nonlinear Extreme Learning Machine for Spectral-Spatial Classification of Hyperspectral Images
Abstract
:1. Introduction
2. Materials and Methods
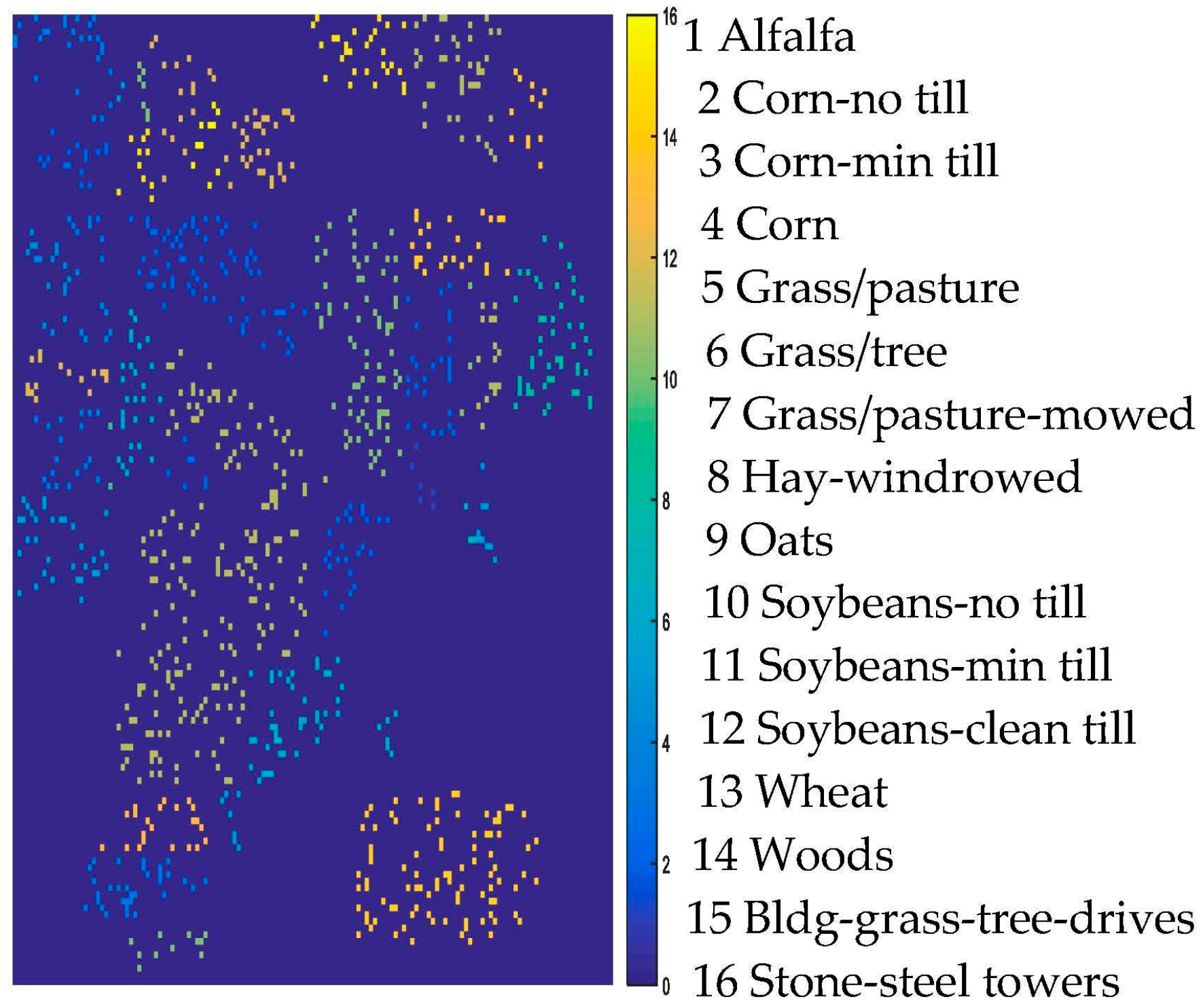
2.1. HSI Data Set
2.2. Normalization
2.3. Linear ELM
2.4. Nonlinear ELM
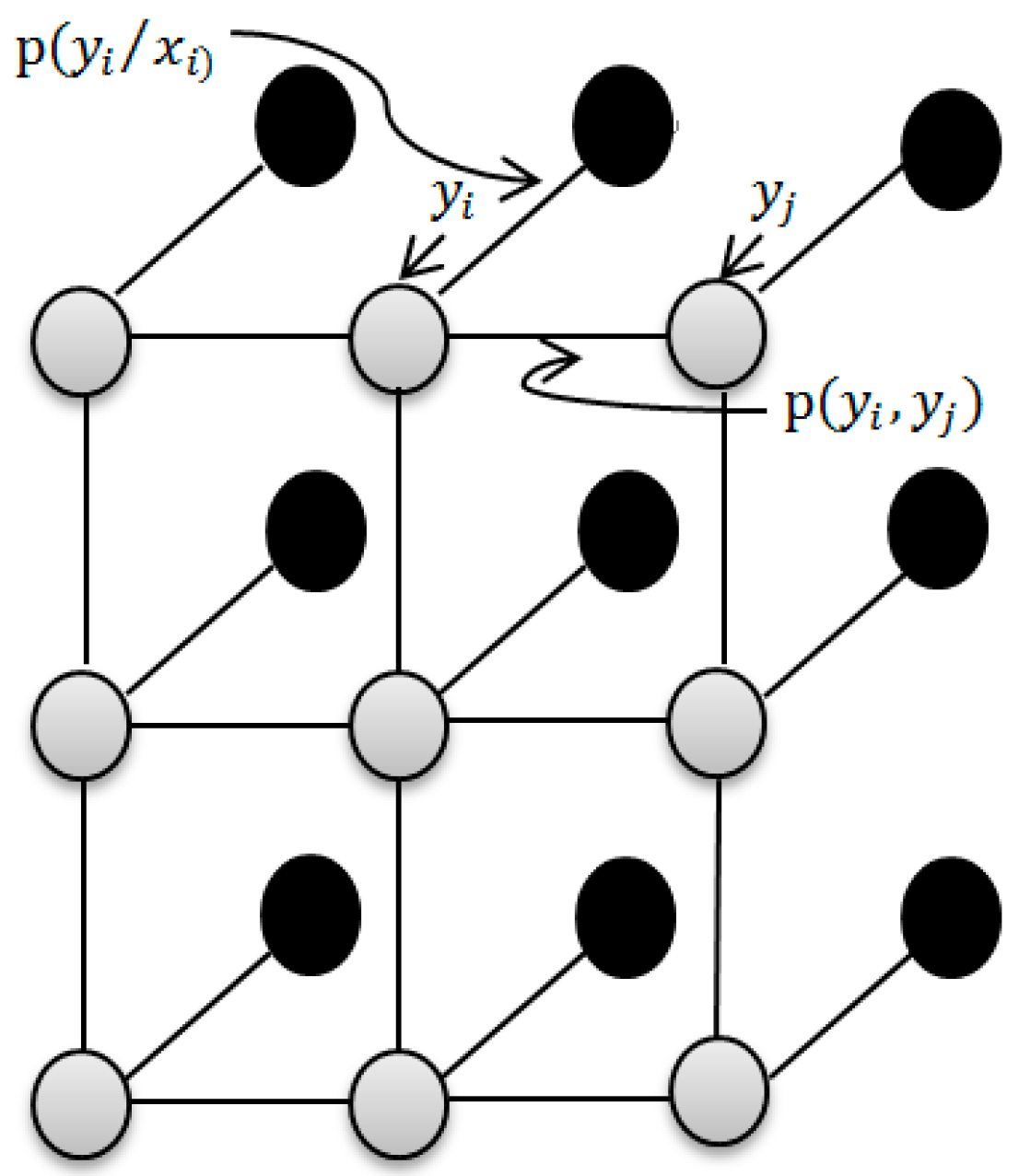
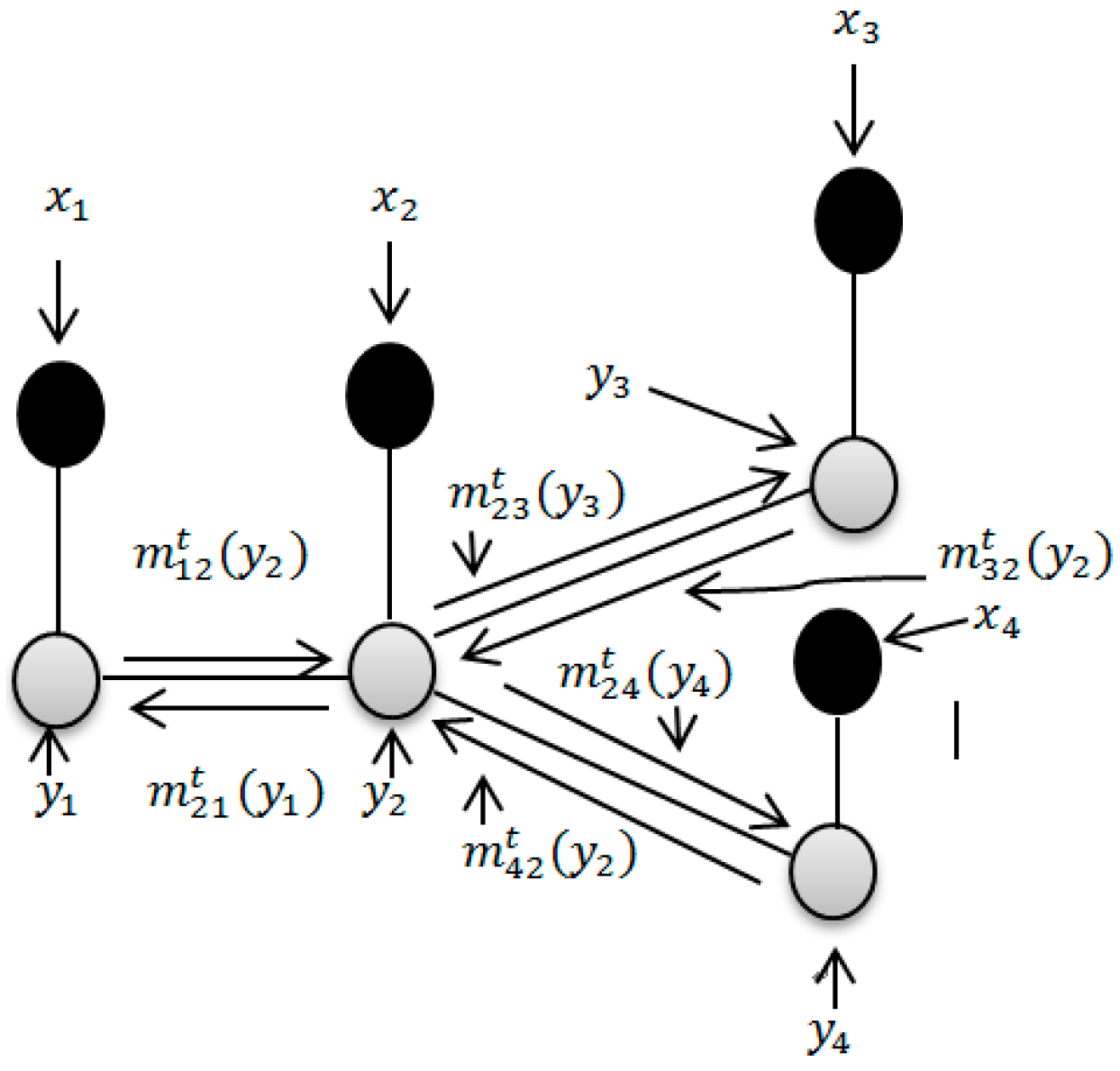
2.5. Using LBP Based Spatial Information to Improve the Classification Accuracy
| Algorithm 1 Spectral-Spatial Classification for HSI Based on LELM and ILBP |
| Input X: the HSI image; : training samples; : test samples; : The desired output of training sample; L: number of hidden node of ELM; g(): activation function of hidden layer of ELM. |
| (1) Normalization: Let , . |
| (2) LELM training: Step 1: Randomly generate the input weights, , and bias, . Step 2: Calculate the hidden layer of the output matrix: |
| Output of LELM: Calculate the hidden layer matrix of the test samples: . Obtain the output result of LELM: . |
(3) Spatial Classification by ILBP:
|
3. Results and Discussions
3.1. Parameter Settings
3.2. Impact of Parameters L and
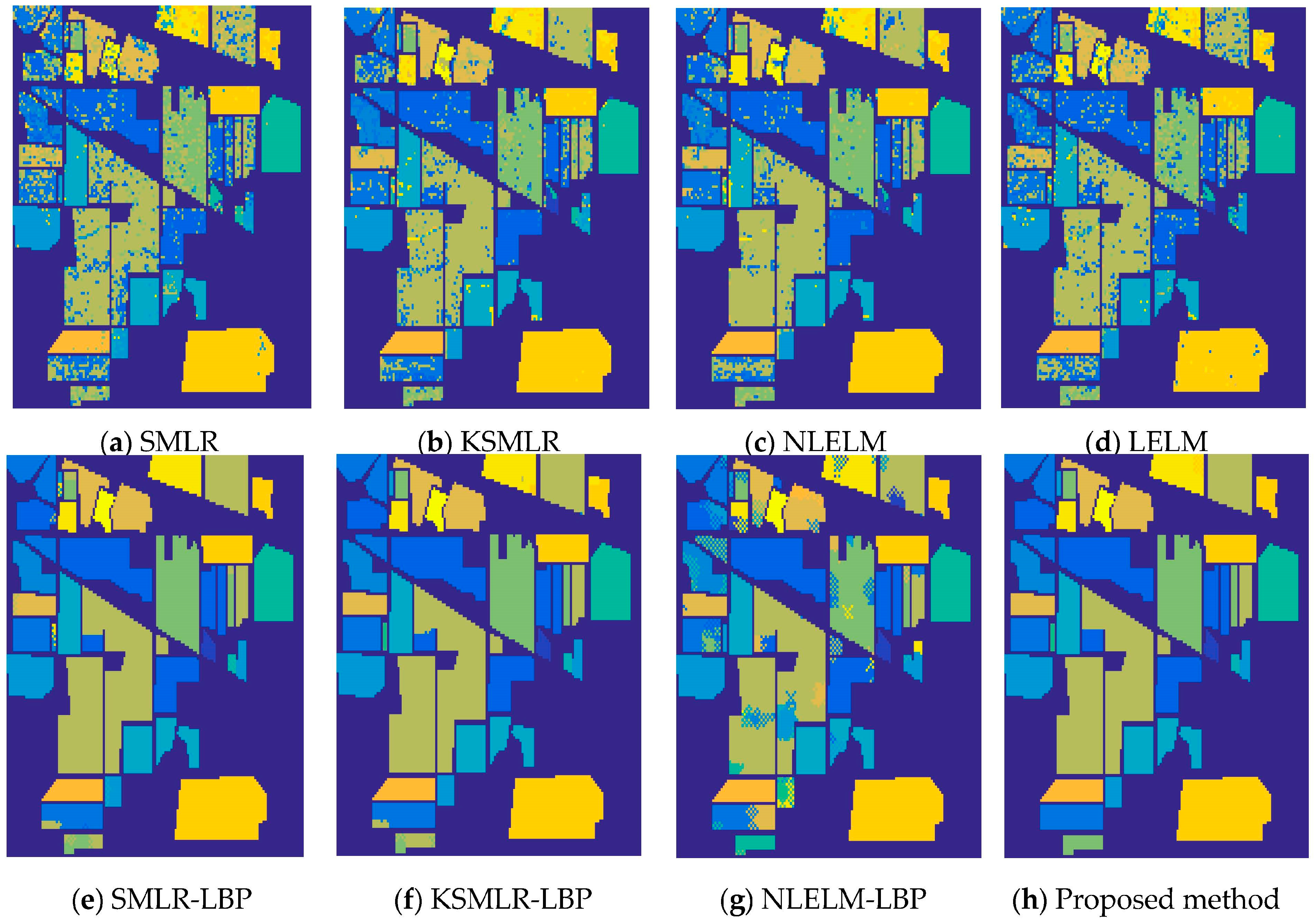
3.3. The Experiment Resutls and Analysis
3.4. The Experiment Resutls and Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhou, Y.; Peng, J.; Chen, C.L.P. Dimension reduction using spatial and spectral regularized local discriminant embedding for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1082–1095. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.; Plaza, A. Spectral–spatial classification of hyperspectral data using loopy belief propagation and active learning. IEEE Trans. Geosci. Remote Sens. 2013, 51, 844–856. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Zheng, J.; Zhao, H.; Qing, C.; Yang, Z.; Du, P.; Marshall, S. Novel segmented stacked autoencoder for effective dimensionality reduction and feature extraction in hyperspectral imaging. Neurocomputing 2016, 185, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Zabalza, Z.; Marshall, S.; Zheng, J. Effective feature extraction and data reduction with hyperspectral imaging in remote sensing. IEEE Signal Process. Mag. 2014, 31, 149–154. [Google Scholar] [CrossRef]
- Qiao, T.; Ren, J.; Wang, X.; Zabalza, J.; Sun, M.; Zhao, H.; Li, S.; Benediktsson, J.A.; Dai, Q.; Marshall, S. Effective denoising and classification of hyperspectral images using curvelet transform and singular spectrum analysis. IEEE Trans. Geosci. Remote Sens. 2017, 55, 119–133. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Zheng, J.; Han, J.; Zhao, H.; Li, S.; Marshall, S. Novel two dimensional singular spectrum analysis for effective feature extraction and data classification in hyperspectral imaging. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4418–4433. [Google Scholar] [CrossRef]
- Qiao, T.; Ren, J.; Craigie, C.; Zabalza, Z.; Maltin, C.; Marshall, S. Singular spectrum analysis for improving hyperspectral imaging based beef eating quality evaluation. Comput. Electron. Agric. 2015, 115, 21–25. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Wang, Z.; Zhao, H.; Wang, J.; Marshall, S. Fast implementation of singular spectrum analysis for effective feature extraction in hyperspectral imaging. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2845–2853. [Google Scholar] [CrossRef]
- Zabalza, J.; Ren, J.; Ren, J.; Liu, Z.; Marshall, S. Structured covaciance principle component analysis for real-time onsite feature extraction and dimensionality reduction in hyperspectral imaging. Appl. Opt. 2014, 53, 4440–4449. [Google Scholar] [CrossRef] [PubMed]
- Zabalza, J.; Ren, J.; Yang, M.; Zhang, Y.; Wang, J.; Marshall, S.; Han, J. Novel Folded-PCA for Improved Feature Extraction and Data Reduction with Hyperspectral Imaging and SAR in Remote Sensing. ISPRS J. Photogramm. Remote Sens. 2014, 93, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Li, S.; Duan, W.; Ren, J.; Benediktsson, J. Classification of hyperspectral images by exploiting spectral-spatial information of superpixel via multiple kernels. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6663–6674. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, F.; Yuan, Y. A study on effectiveness of extreme learning machine. Neurocomputing 2011, 74, 2483–2490. [Google Scholar] [CrossRef]
- Rong, H.J.; Ong, Y.S.; Tan, A.H.; Zhu, Z. A fast pruned-extreme learning machine for classification problem. Neurocomputing 2008, 72, 359–366. [Google Scholar] [CrossRef]
- Huang, G.B.; Ding, X.; Zhou, H. Optimization method based extreme learning machine for classification. Neurocomputing 2010, 74, 155–163. [Google Scholar] [CrossRef]
- Samat, A.; Du, P.; Liu, S.; Li, J.; Cheng, L. Ensemble Extreme Learning Machines for Hyperspectral Image Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1060–1069. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhou, H.; Ding, X.; Zhang, L. Extreme learning machine for regression and multiclass classification. IEEE Trans. Syst. Man Cybern. B Cybern. 2012, 42, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Huang, G.B.; Wang, D.; Wang, H.; Westover, M.B. Sparse extreme learning machine for classification. IEEE Trans. Cybern. 2014, 44, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Peng, J.; Chen, C.L.P. Extreme learning machine with composite kernels for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2351–2360. [Google Scholar] [CrossRef]
- Chen, C.; Li, W.; Su, H.; Liu, K. Spectral-spatial classification of hyperspectral image based on kernel extreme learning machine. Remote Sens. 2014, 6, 5795–5814. [Google Scholar] [CrossRef]
- Duan, W.; Li, S.; Fang, L. Spectral-spatial hyperspectral image classification using superpixel and extreme learning machines. In Chinese Conference on Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2014; pp. 159–167. [Google Scholar]
- Argüello, F.; Heras, D.B. ELM-based spectral–spatial classification of hyperspectral images using extended morphological profiles and composite feature mappings. Int. J. Remote Sens. 2015, 36, 645–664. [Google Scholar] [CrossRef]
- Yedidia, J.S.; Freeman, W.T.; Weiss, Y. Understanding belief propagation and its generalizations. In Exploring Artificial Intelligence in the New Millennium; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2003; pp. 236–239. [Google Scholar]
- Yedidia, J.S.; Freeman, W.T.; Weiss, Y. Constructing free-energy approximations and generalized belief propagation algorithms. IEEE Trans. Inf. Theory 2005, 51, 2282–2312. [Google Scholar] [CrossRef]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J.C. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE. 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J.; Benediktsson, J.A. SVM-and MRF-based method for accurate classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2010, 7, 736–740. [Google Scholar] [CrossRef]
- Ghamisi, P.; Benediktsson, J.A.; Ulfarsson, M.O. Spectral-spatial classification of hyperspectral images based on hidden Markov random fields. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2565–2574. [Google Scholar] [CrossRef]
- Damodaran, B.B.; Nidamanuri, R.R.; Tarabalka, Y. Dynamic ensemble selection approach for hyperspectral image classification with joint spectral and spatial information. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2405–2417. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Su, H.; Du, Q. Local binary patterns and extreme learning machine for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3681–3693. [Google Scholar] [CrossRef]
- Kumar, S.; Hebert, M. Discriminative random fields. Int. J. Comput. Vis. 2006, 68, 179–201. [Google Scholar] [CrossRef]
- Li, S.Z. Markov Random Field Modeling in Computer Vision; Springer: Berlin/Heidelberg, Germany, 1994. [Google Scholar]
- Borges, J.S.; Marçal, A.R.S.; Bioucas-Dias, J.M. Evaluation of Bayesian hyperspectral image segmentation with a discriminative class learning. In Proceedings of the IEEE International Symposium on Geoscience and Remote Sensing, Barcelona, Spain, 23–28 July 2003; pp. 3810–3813. [Google Scholar]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Semisupervised hyperspectral image segmentation using multinomial logistic regression with active learning. IEEE Trans. Geosci. Remote Sens. 2010, 4298, 4085–4098. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, H.; Pizurica, A. A Robust Sparse Representation Model for Hyperspectral Image Classification. Sensors 2017, 17, 2087. [Google Scholar] [CrossRef] [PubMed]
- Geman, S.; Geman, D. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell. 1984, 6, 721–741. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Hyperspectral image segmentation using a new Bayesian approach with active learning. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3947–3960. [Google Scholar] [CrossRef]
- Sun, L.; Wu, Z.; Liu, J.; Xiao, L.; Wei, Z. Supervised spectral–spatial hyperspectral image classification with weighted Markov random fields. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1490–1503. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.; Figueiredo, M. Logistic Regression via Variable Splitting and Augmented Lagrangian Tools; Technical Report; Instituto Superior Técnico: Lisboa, Portugal, 2009. [Google Scholar]
- Li, H.; Li, C.; Zhang, C.; Liu, Z.; Liu, C. Hyperspectral Image Classification with Spatial Filtering and ℓ2,1 Norm. Sensors 2017, 17, 314. [Google Scholar]
- Mura, M.; Benediktsson, J.A.; Waske, B.; Bruzzone, L. Morphological attribute profiles for the analysis of very high resolution images. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3747–3762. [Google Scholar] [CrossRef]


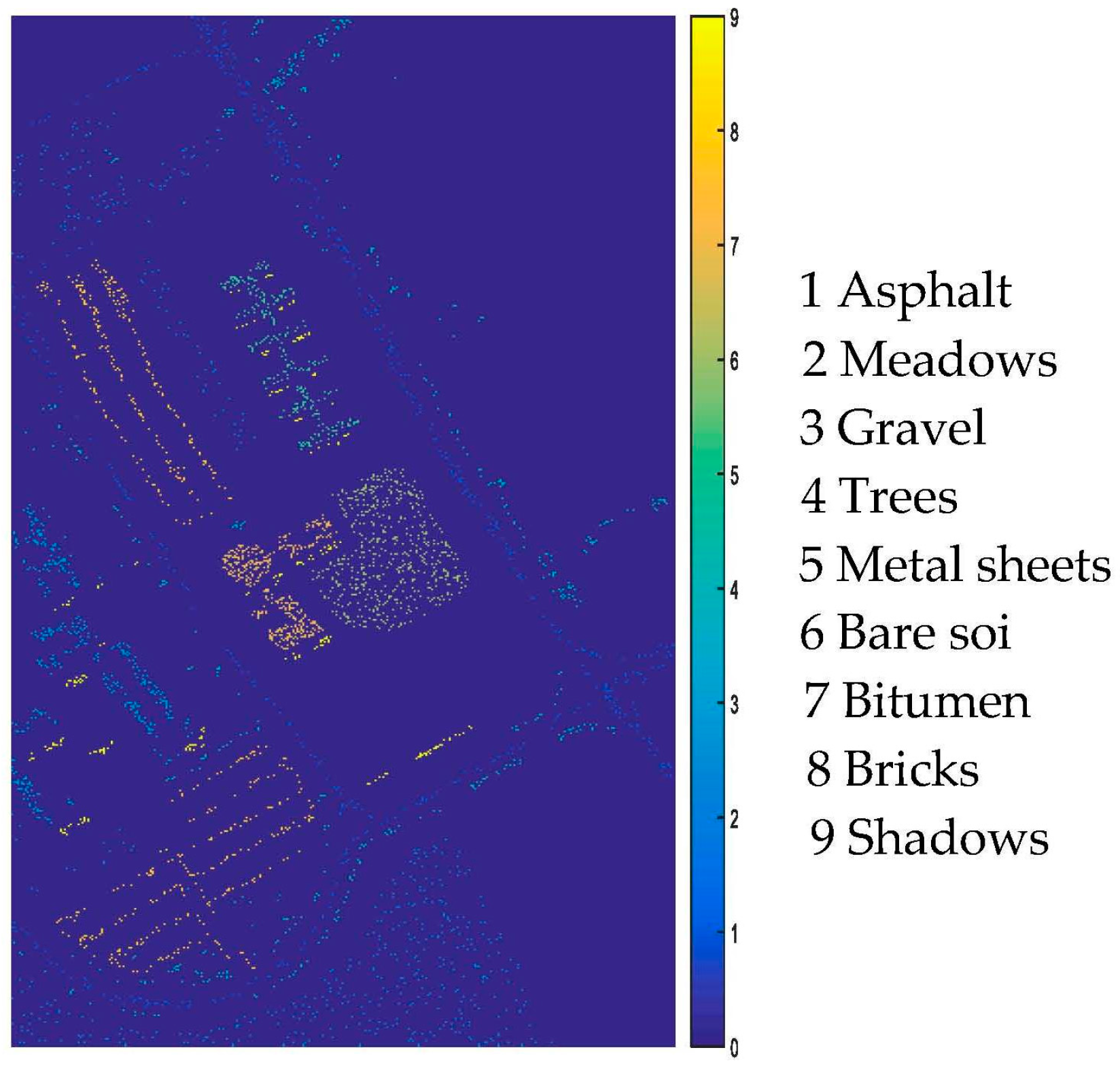
| Indian Pines | Pavia University | |||||||
|---|---|---|---|---|---|---|---|---|
| Class | Train | Test | Class | Train | Test | Class | Train | Test |
| Alfalfa | 6 | 54 | Oats | 2 | 20 | Asphalt | 548 | 6631 |
| Corn-no till | 144 | 1434 | Soybeans-no till | 97 | 968 | Meadows | 548 | 18,649 |
| Corn-min till | 84 | 834 | Soybeans-min till | 247 | 2468 | Gravel | 392 | 2099 |
| Corn | 24 | 234 | Soybeans-clean till | 62 | 614 | Trees | 524 | 3064 |
| Grass/pasture | 50 | 497 | Wheat | 22 | 212 | Metal sheets | 265 | 1345 |
| Grass/tree | 75 | 747 | Woods | 130 | 1294 | Bare soil | 532 | 5029 |
| Grass/pasture-mowed | 3 | 26 | Bldg-grass-tree-drives | 38 | 380 | Bitumen | 375 | 1330 |
| Hay-windrowed | 49 | 489 | Stone-steel towers | 10 | 95 | Bricks | 514 | 3682 |
| Total | 1043 | 10366 | Shadows | 231 | 947 | |||
| Total | 3921 | 42,776 | ||||||
| Class | SMLR | KSMLR | LELM | NLELM | SMLR-LBP | KSMLR-LBP | NLELM-LBP | PROPOSED METHOD |
|---|---|---|---|---|---|---|---|---|
| Alfalfa | 30.52 | 74.26 | 35.37 | 71.11 | 97.78 | 100 | 90.37 | 100.00 |
| Corn-no till | 75.87 | 82.49 | 79.27 | 85.82 | 99.02 | 99.40 | 85.68 | 99.68 |
| Corn-min till | 51.35 | 70.86 | 58.26 | 72.58 | 92.55 | 97.35 | 68.79 | 99.22 |
| Corn | 37.35 | 68.68 | 43.29 | 69.10 | 99.27 | 95.00 | 77.44 | 100.00 |
| Grass/pasture | 86.82 | 89.46 | 89.76 | 93.64 | 97.36 | 98.23 | 93.64 | 99.28 |
| Grass/tree | 94.28 | 96.37 | 96.32 | 97.39 | 100.00 | 100.00 | 95.70 | 100.00 |
| Grass/pasture-mowed | 6.92 | 45.00 | 11.54 | 70.38 | 71.92 | 91.54 | 45.00 | 95.38 |
| Hay-windrowed | 99.37 | 98.51 | 99.57 | 99.04 | 100.00 | 100 | 98.73 | 100.00 |
| Oats | 5 | 38.50 | 11.50 | 63.50 | 16.50 | 100 | 48.00 | 100.00 |
| Soybeans-no till | 61.03 | 74.91 | 66.69 | 80.79 | 96.27 | 96.34 | 80.74 | 99.23 |
| Soybeans-min till | 74.46 | 84.51 | 80.23 | 87.66 | 99.96 | 99.91 | 90.41 | 99.93 |
| Soybeans-clean till | 68.96 | 82.20 | 72.98 | 84.98 | 98.50 | 100 | 82.85 | 100.00 |
| Wheat | 96.75 | 99.15 | 99.39 | 98.96 | 100.00 | 100 | 98.77 | 100.00 |
| Woods | 95.04 | 95.20 | 95.65 | 96.51 | 100.00 | 99.69 | 97.26 | 100.00 |
| Bldg-grass-tree-drives | 67.13 | 73.05 | 64.08 | 70.45 | 95.47 | 99.50 | 83.53 | 99.89 |
| Stone-steel towers | 69.26 | 70.32 | 70.42 | 77.05 | 99.58 | 98.63 | 98.63 | 99.89 |
| OA | 75.76 | 84.34 | 79.43 | 86.93 | 98.26 | 99.05 | 87.95 | 99.75 |
| AA | 63.66 | 77.72 | 67.15 | 82.44 | 91.51 | 98.47 | 83.47 | 99.53 |
| k | 72.22 | 82.09 | 76.38 | 85.06 | 98.02 | 98.92 | 86.36 | 99.72 |
| Execution Time (seconds) | 0.02 | 0.41 | 0.19 | 0.31 | 38.74 | 40.70 | 39.59 | 38.95 |
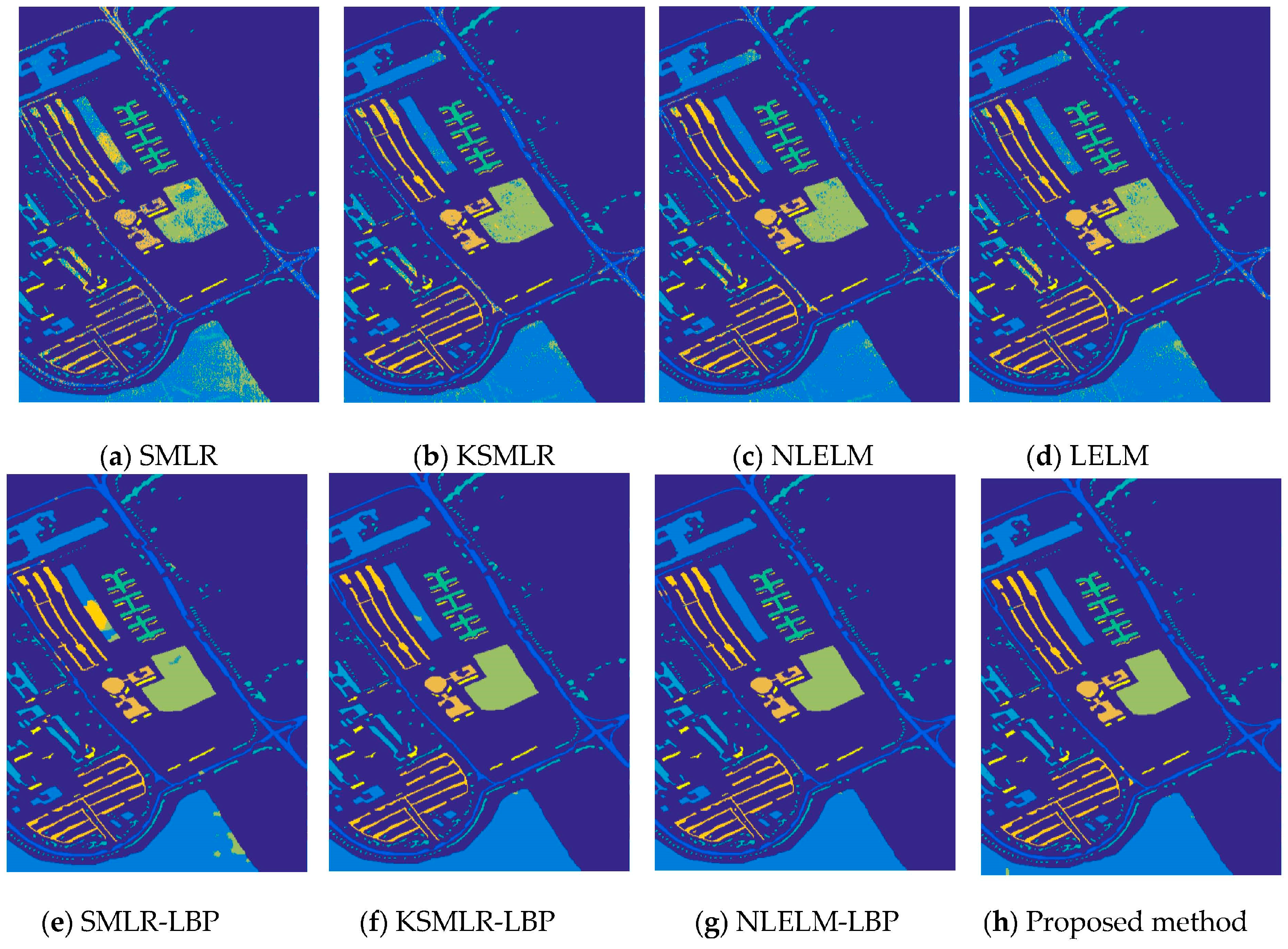
| Class | SMLR | KSMLR | LELM | NLELM | SMLR-LBP | KSMLR-LBP | NLELM-LBP | PROPOSED METHOD |
|---|---|---|---|---|---|---|---|---|
| Asphalt | 72.27 | 89.43 | 85.27 | 88.82 | 98.62 | 99.63 | 99.49 | 99.63 |
| Meadows | 79.08 | 94.16 | 92.17 | 94.61 | 93.70 | 99.34 | 99.88 | 99.83 |
| Gravel | 71.99 | 85.08 | 78.06 | 87.41 | 99.14 | 99.64 | 99.92 | 99.83 |
| Trees | 94.90 | 97.92 | 97.38 | 98.16 | 99.27 | 99.86 | 98.54 | 99.64 |
| Metal sheets | 99.58 | 99.34 | 98.85 | 99.39 | 100.00 | 100.00 | 100.00 | 100.00 |
| Bare soil | 74.26 | 94.77 | 93.90 | 95.43 | 99.93 | 100.00 | 100.00 | 100.00 |
| Bitumen | 78.66 | 93.82 | 93.69 | 95.34 | 100.00 | 100.00 | 100.00 | 100.00 |
| Bricks | 73.37 | 87.52 | 90.05 | 90.94 | 99.93 | 99.63 | 99.85 | 100.00 |
| Shadows | 96.88 | 99.61 | 99.70 | 99.97 | 99.89 | 99.87 | 94.14 | 99.89 |
| OA | 78.78 | 93.00 | 91.23 | 93.94 | 96.93 | 99.59 | 99.62 | 99.83 |
| AA | 82.33 | 93.49 | 92.12 | 94.56 | 98.94 | 99.77 | 99.09 | 99.87 |
| k | 72.73 | 90.82 | 88.54 | 92.04 | 95.98 | 99.46 | 99.49 | 99.78 |
| Execution Time (seconds) | 0.19 | 4.40 | 0.48 | 3.83 | 1193.7 | 1237.1 | 5288.6 | 1201.2 |
| Datasets | Index | EMP-ELM | S-ELM | G-ELM | PROPOSED METHOD |
|---|---|---|---|---|---|
| Indian Pines data set with 10% training samples | OA | - | 97.78 | 99.08 | 99.75 |
| AA | - | 97.10 | 98.68 | 99.53 | |
| k | - | 97 | 98.95 | 99.72 | |
| Pavia University data set with 9% training samples | OA | 99.65 | - | - | 99.83 |
| AA | 99.60 | - | - | 99.87 | |
| k | 99.52 | - | - | 99.78 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, F.; Yang, Z.; Ren, J.; Jiang, M.; Ling, W.-K. Linear vs. Nonlinear Extreme Learning Machine for Spectral-Spatial Classification of Hyperspectral Images. Sensors 2017, 17, 2603. https://doi.org/10.3390/s17112603
Cao F, Yang Z, Ren J, Jiang M, Ling W-K. Linear vs. Nonlinear Extreme Learning Machine for Spectral-Spatial Classification of Hyperspectral Images. Sensors. 2017; 17(11):2603. https://doi.org/10.3390/s17112603
Chicago/Turabian StyleCao, Faxian, Zhijing Yang, Jinchang Ren, Mengying Jiang, and Wing-Kuen Ling. 2017. "Linear vs. Nonlinear Extreme Learning Machine for Spectral-Spatial Classification of Hyperspectral Images" Sensors 17, no. 11: 2603. https://doi.org/10.3390/s17112603





