Estimation of Electrically-Evoked Knee Torque from Mechanomyography Using Support Vector Regression
Abstract
:1. Introduction
2. Materials and Methods
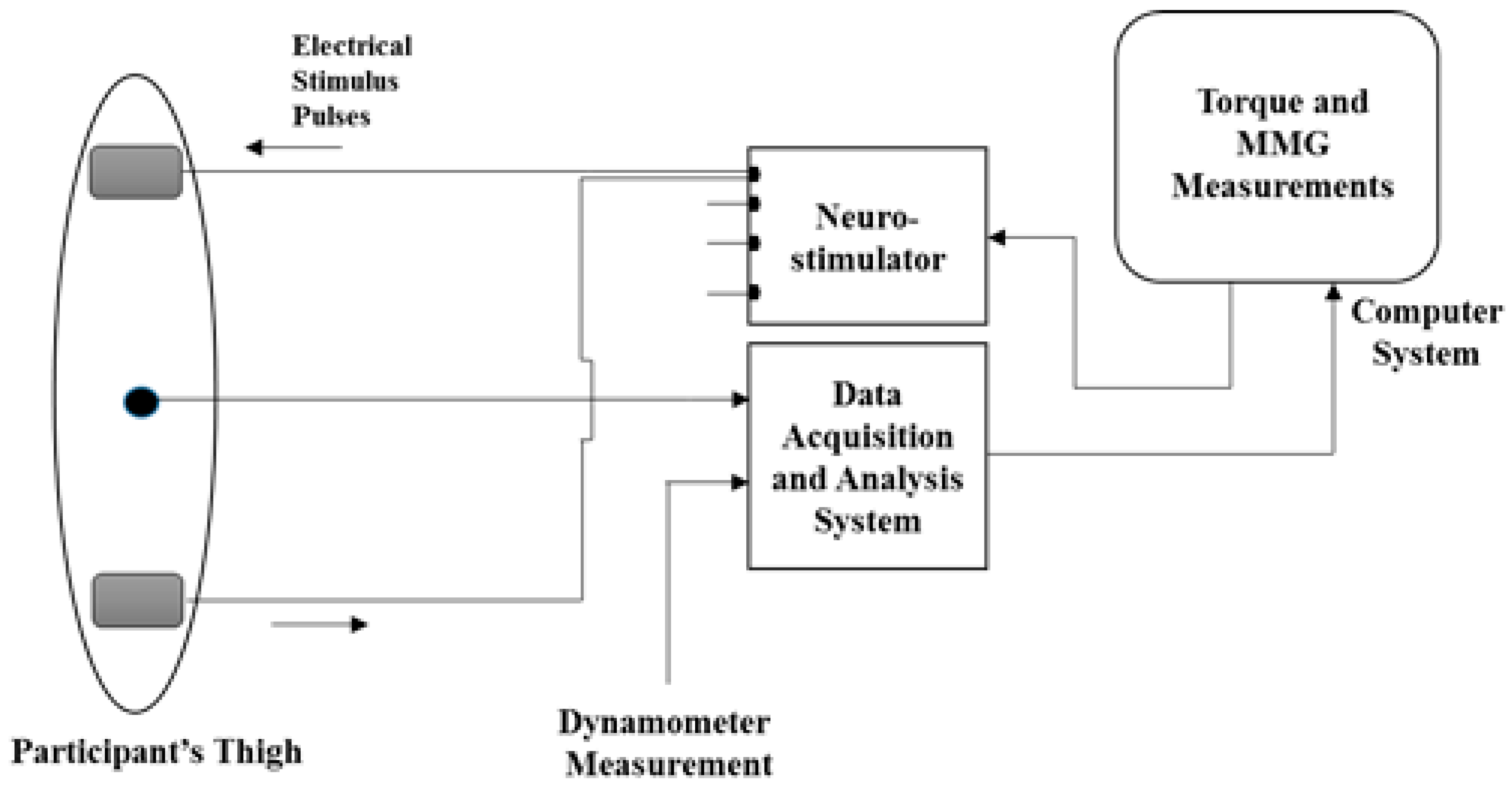
2.1. Experimental Protocol
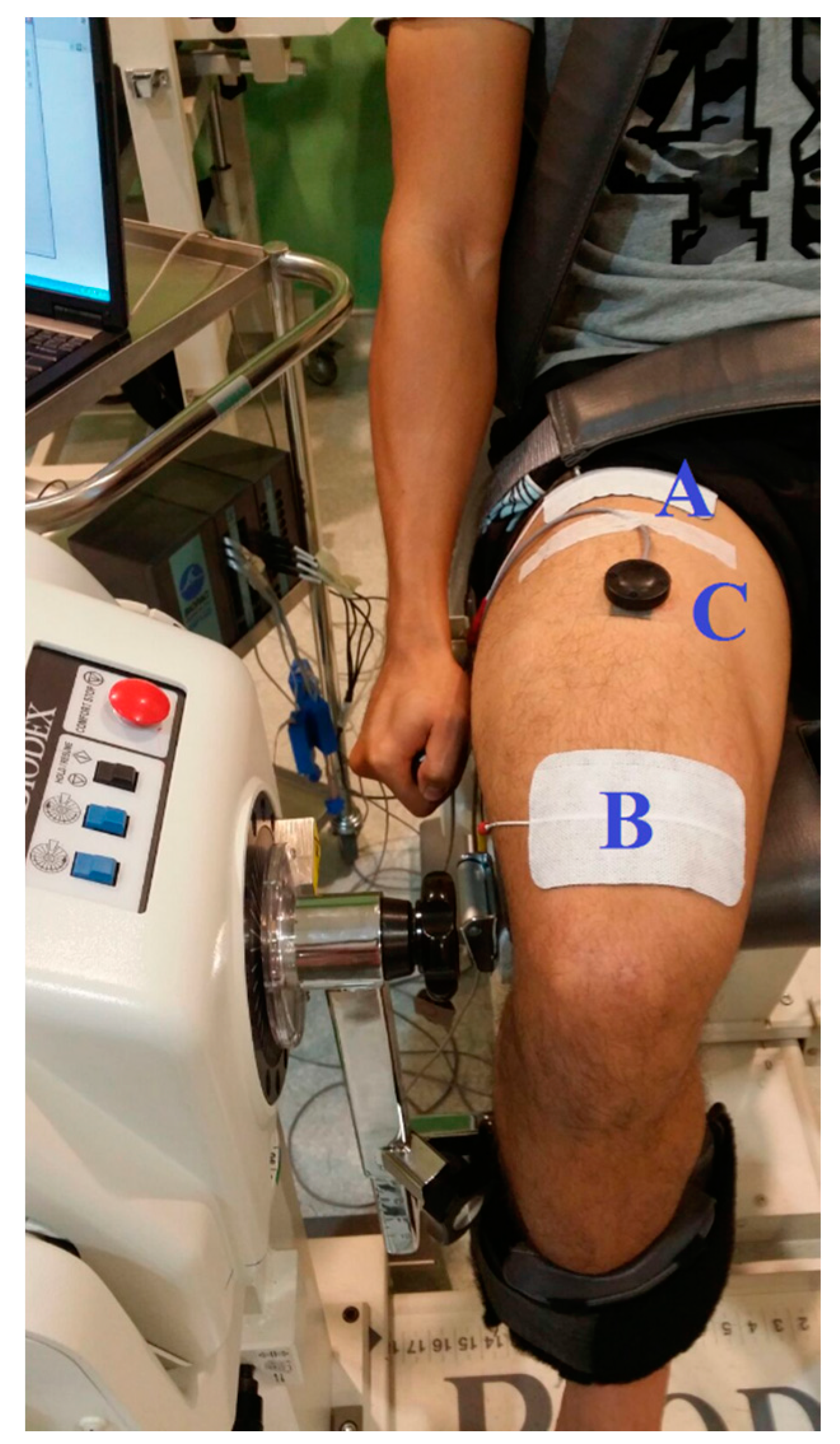
2.1.1. NMES-Evoked Muscle Contractions and Knee Torque Measurements
2.1.2. MMG Acquisition and Processing
2.2. Support Vector Regression Modelling Approach
2.2.1. Model Development
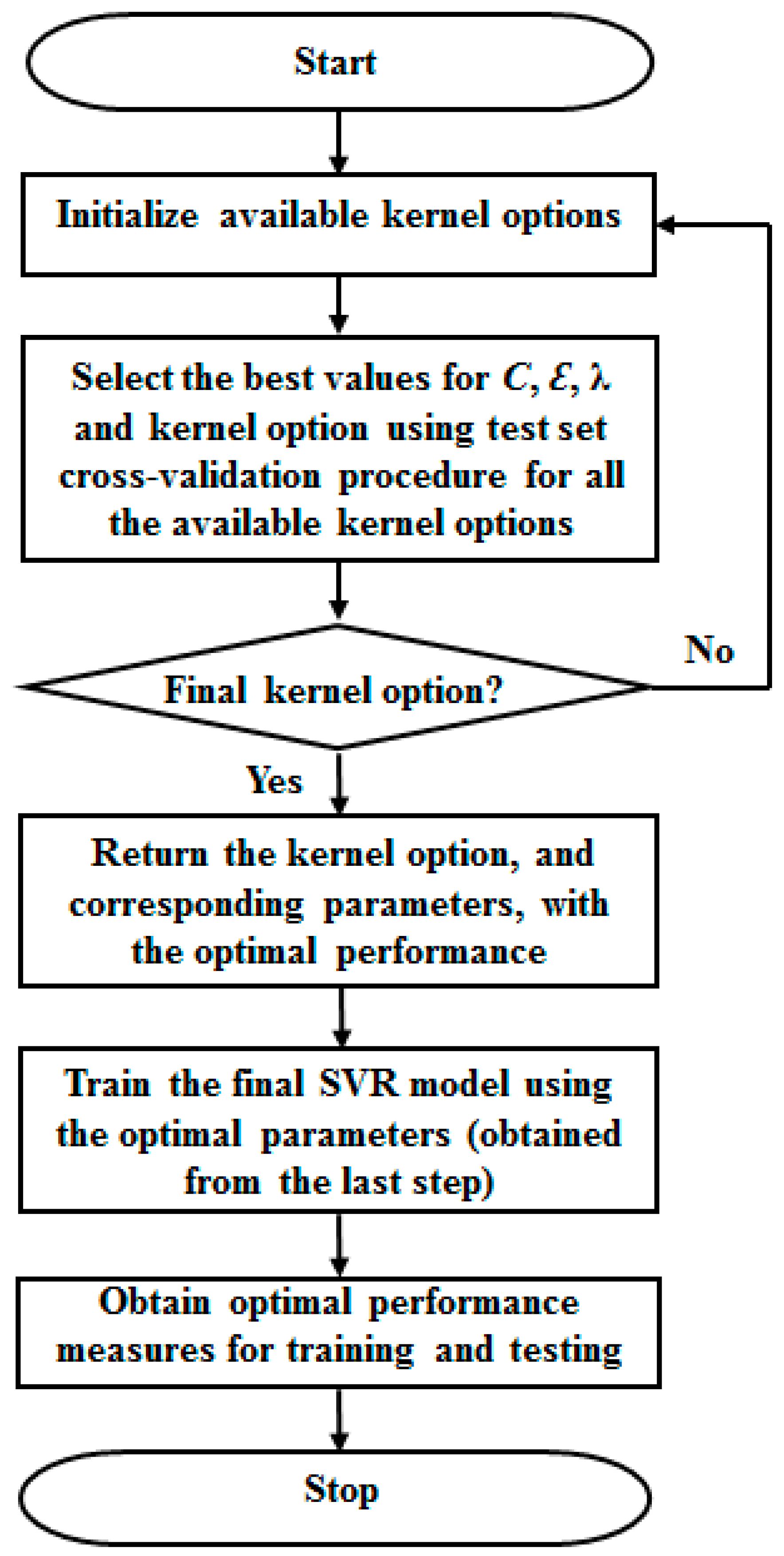
2.2.2. Optimal Parameters Search Approach
| Algorithm 1. Optimal parameter search algorithm |
| , , , |
| {Performance measure for the present parameters combination} |
| end |
| end |
| end |
2.2.3. Model Statistical Performance Criteria
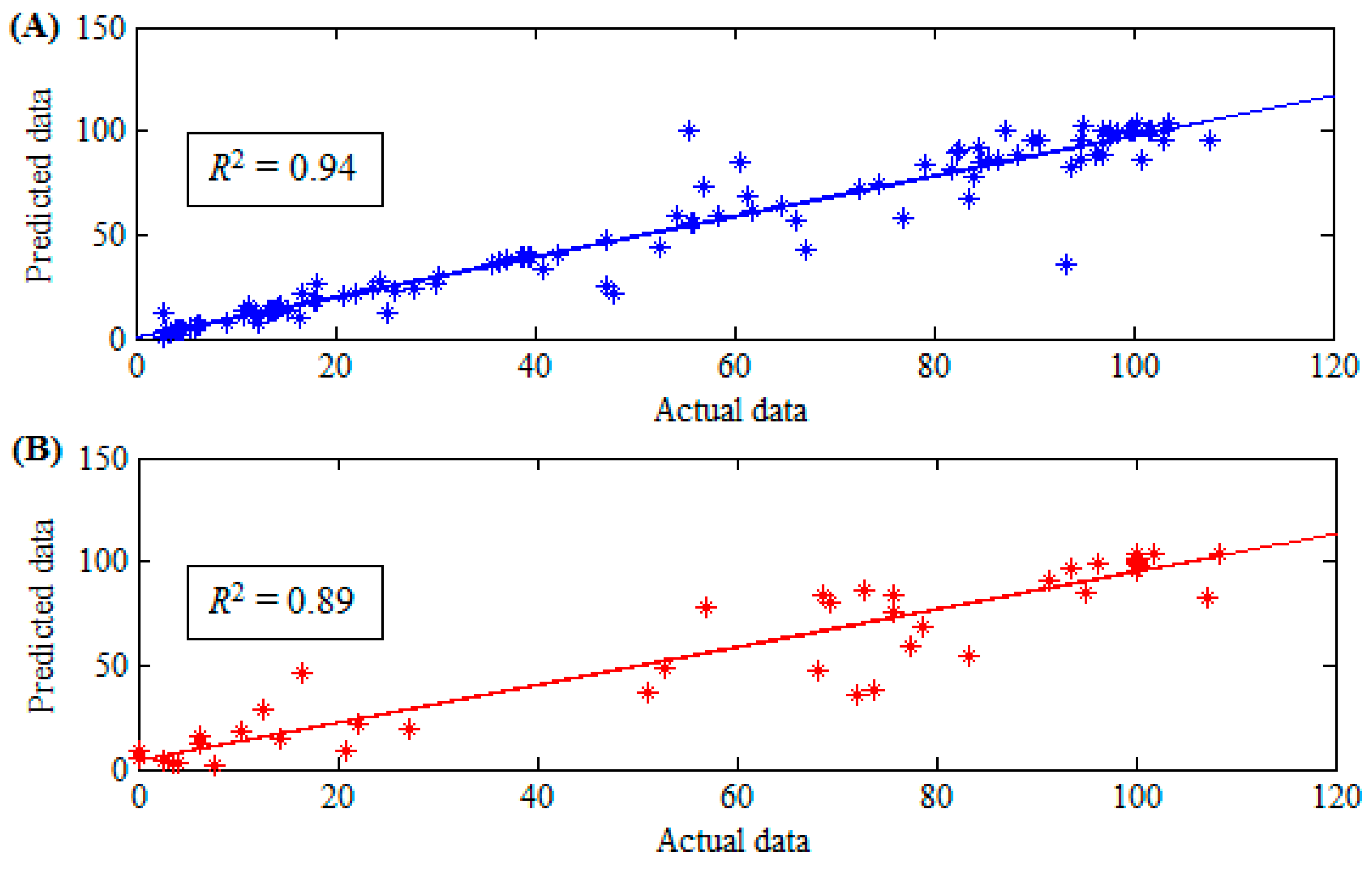
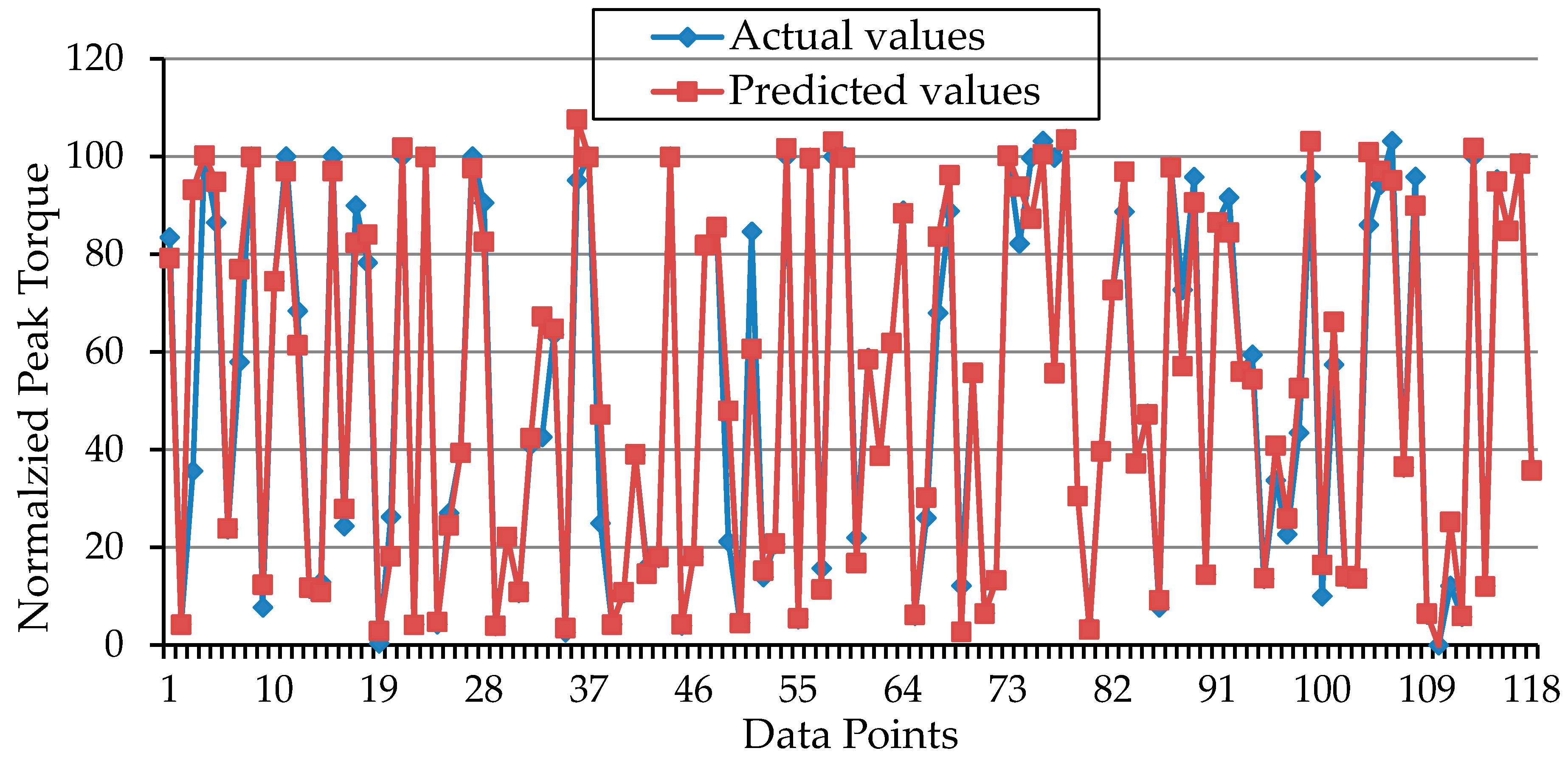
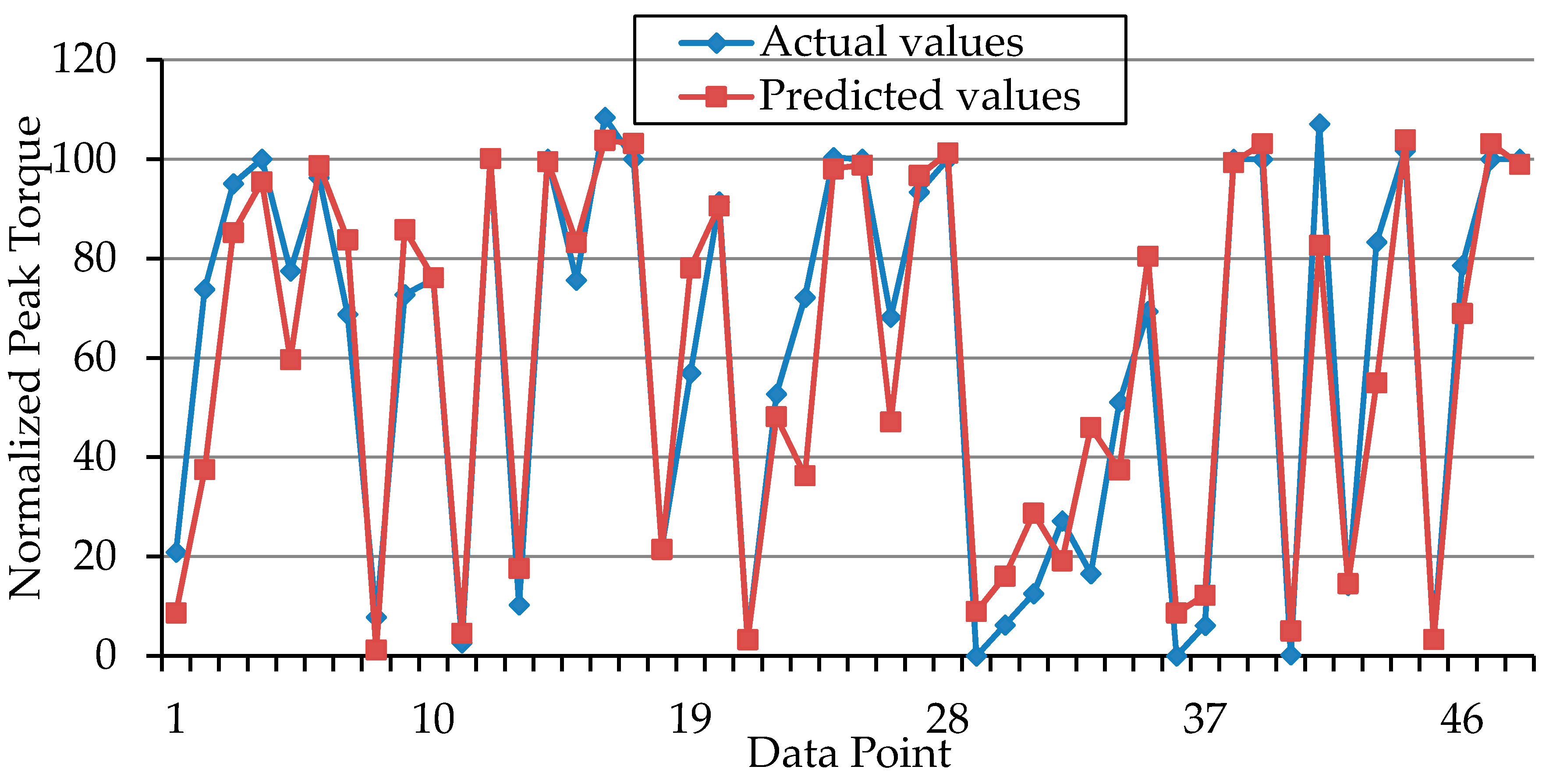
3. Results and Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NMES | Neuromuscular Electrical Stimulation |
| MMG | Mechanomyography |
| SVR | Support Vector Regression |
| SVM | Support Vector Machine |
| RMSE | Root Mean Square Error |
| PT | Peak Torque |
| RF | Rectus Femoris |
| RMS | Root Mean Square |
| PTP | Peak to Peak |
References
- Brocherie, F.; Babault, N.; Cometti, G.; Maffiuletti, N.; Chatard, J.-C. Electrostimulation training effects on the physical performance of ice hockey players. Med. Sci. Sports Exerc. 2005, 37, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.G.; Bennett, M.J.; Hieb, M.A.; Hollar, A.C.; Roe, A.A. Strength response in human quadriceps femoris muscle during 2 neuromuscular electrical stimulation programs. J. Orthop. Sports Phy. Ther. 2003, 33, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Braz, G.P.; Russold, M.; Davis, G.M. Functional electrical stimulation control of standing and stepping after spinal cord injury: A review of technical characteristics. Neuromodul. Technol. Neural Interface 2009, 12, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Deley, G.; Denuziller, J.; Babault, N.; Taylor, J.A. Effects of electrical stimulation pattern on quadriceps isometric force and fatigue in individuals with spinal cord injury. Muscle Nerve 2015, 52, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Popović, D.B. Advances in functional electrical stimulation (fes). J. Electromyogr. Kinesiol. 2014, 24, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Orizio, C. Muscle sound: Bases for the introduction of a mechanomyographic signal in muscle studies. Crit. Rev. Biomed. Eng. 1993, 21, 201–243. [Google Scholar] [PubMed]
- Ibitoye, M.O.; Hamzaid, N.A.; Zuniga, J.M.; Abdul Wahab, A.K. Mechanomyography and muscle function assessment: A review of current state and prospects. Clin. Biomech. 2014, 29, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Farina, D.; Li, X.; Madeleine, P. Motor unit acceleration maps and interference mechanomyographic distribution. J. Biomech. 2008, 41, 2843–2849. [Google Scholar] [CrossRef] [PubMed]
- Watakabe, M.; Itoh, Y.; Mita, K.; Akataki, K. Technical aspects of mechnomyography recording with piezoelectric contact sensor. Med. Biol. Eng. Comput. 1998, 36, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Yuan-Ting, Z.; Frank, C.B.; Rangayyan, R.M.; Bell, G.D. A comparative study of simultaneous vibromyography and electromyography with active human quadriceps. IEEE Trans. Biomed. Eng. 1992, 39, 1045–1052. [Google Scholar]
- Matheson, G.O.; Maffey-Ward, L.; Mooney, M.; Ladly, K.; Fung, T.; Zhang, Y.T. Vibromyography as a quantitative measure of muscle force production. Scand. J. Rehabil. Med. 1997, 29, 29–35. [Google Scholar] [PubMed]
- Beck, T.W.; Housh, T.J.; Johnson, G.O.; Weir, J.P.; Cramer, J.T.; Coburn, J.W.; Malek, M.H. Mechanomyographic amplitude and mean power frequency versus torque relationships during isokinetic and isometric muscle actions of the biceps brachii. J. Electromyogr. Kinesiol. 2004, 14, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Youn, W.; Kim, J. Estimation of elbow flexion force during isometric muscle contraction from mechanomyography and electromyography. Med. Biol. Eng. Comput. 2010, 48, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Alves, N.; Chau, T. The design and testing of a novel mechanomyogram-driven switch controlled by small eyebrow movements. J. NeuroEng. Rehabil. 2010, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ibitoye, M.O.; Hamzaid, N.A.; Zuniga, J.M.; Hasnan, N.; Wahab, A.K.A. Mechanomyographic parameter extraction methods: An appraisal for clinical applications. Sensors 2014, 14, 22940–22970. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Heim, W.; Chau, T. A self-contained, mechanomyography-driven externally powered prosthesis. Arch. Phys. Med. Rehabil. 2005, 86, 2066–2070. [Google Scholar] [CrossRef] [PubMed]
- Orizio, C.; Diemont, B.; Esposito, F.; Alfonsi, E.; Parrinello, G.; Moglia, A.; Veicsteinas, A. Surface mechanomyogram reflects the changes in the mechanical properties of muscle at fatigue. Eur. J. Appl. Physiol. Occup. Physiol. 1999, 80, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Hong-Bo, X.; Yong-Ping, Z.; Jing-Yi, G. Classification of the mechanomyogram signal using a wavelet packet transform and singular value decomposition for multifunction prosthesis control. Physiol. Meas. 2009, 30, 441–457. [Google Scholar]
- Barry, D.T.; Leonard, J.A.; Gitter, A.J.; Ball, R.D. Acoustic myography as a control signal for an externally powered prosthesis. Arch. Phys. Med. Rehabil. 1986, 67, 267–269. [Google Scholar] [PubMed]
- Gobbo, M.; Cè, E.; Diemont, B.; Esposito, F.; Orizio, C. Torque and surface mechanomyogram parallel reduction during fatiguing stimulation in human muscles. Eur. J. Appl. Physiol. 2006, 97, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.-B.; Zheng, Y.-P.; Guo, J.-Y.; Chen, X.; Shi, J. Estimation of wrist angle from sonomyography using support vector machine and artificial neural network models. Med. Eng. Phys. 2009, 31, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Youn, W.; Kim, J. Feasibility of using an artificial neural network model to estimate the elbow flexion force from mechanomyography. J. Neurosci. Methods 2011, 194, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Shamshirband, S.; Petković, D.; Amini, A.; Anuar, N.B.; Nikolić, V.; Ćojbašić, Ž.; Mat Kiah, M.L.; Gani, A. Support vector regression methodology for wind turbine reaction torque prediction with power-split hydrostatic continuous variable transmission. Energy 2014, 67, 623–630. [Google Scholar] [CrossRef]
- Vapnik, V.; Golowich, S.; Smola, A. Support vector method for function approximation, regression estimation, and signal processing. In Advances in Neural Information Processing Systems; Mozer, M.C., Jordan, M.I., Petsche, T., Eds.; The MIT Press: Cambridge, MA, USA, 1997; Volume 9, pp. 281–287. [Google Scholar]
- Yang, H.; Huang, K.; King, I.; Lyu, M.R. Localized support vector regression for time series prediction. Neurocomputing 2009, 72, 2659–2669. [Google Scholar] [CrossRef]
- Jiang, H.; He, W. Grey relational grade in local support vector regression for financial time series prediction. Expert Syst. Appl. 2012, 39, 2256–2262. [Google Scholar] [CrossRef]
- Yu, W.; Liu, T.; Valdez, R.; Gwinn, M.; Khoury, M. Application of support vector machine modeling for prediction of common diseases: The case of diabetes and pre-diabetes. BMC Med. Inform. Decis. Mak. 2010, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Ziai, A.; Menon, C. Comparison of regression models for estimation of isometric wrist joint torques using surface electromyography. J. NeuroEng. Rehabil. 2011, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Güler, N.; Koçer, S. Use of support vector machines and neural network in diagnosis of neuromuscular disorders. J. Med. Syst. 2005, 29, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.E.; Weir, J.P. Asep procedures recommendation i: Accurate assessment of muscular strength and power. J. Exerc. Physiol. 2001, 4, 1–21. [Google Scholar]
- Bickel, C.S.; Slade, J.M.; VanHiel, L.R.; Warren, G.L.; Dudley, G.A. Variable-frequency-train stimulation of skeletal muscle after spinal cord injury. J. Rehabil. Res. Dev. 2004, 41, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Orizio, C.; Solomonow, M.; Baratta, R.; Veicsteinas, A. Influence of motor units recruitment and firing rate on the soundmyogram and emg characteristics in cat gastrocnemius. J. Electromyogr. Kinesiol. 1992, 2, 232–241. [Google Scholar] [CrossRef]
- Adams, G.R.; Harris, R.T.; Woodard, D.; Dudley, G.A. Mapping of electrical muscle stimulation using mri. J. Appl. Physiol. 1993, 74, 532–537. [Google Scholar] [PubMed]
- Selkowitz, D.M. Improvement in isometric strength of the quadriceps femoris muscle after training with electrical stimulation. Phys. Ther. 1985, 65, 186–196. [Google Scholar] [PubMed]
- Babault, N.; Pousson, M.; Ballay, Y.; Van Hoecke, J. Activation of human quadriceps femoris during isometric, concentric, and eccentric contractions. J. Appl. Physiol. 2001, 91, 2628–2634. [Google Scholar] [PubMed]
- Levin, O.; Mizrahi, J.; Isakov, E. Transcutaneous fes of the paralyzed quadriceps: Is knee torque affected by unintended activation of the hamstrings? J. Electromyogr. Kinesiol. 2000, 10, 47–58. [Google Scholar] [CrossRef]
- Ebersole, K.T.; Housh, T.J.; Johnson, G.O.; Evetovich, T.K.; Smith, D.B.; Perry, S.R. The effect of leg flexion angle on the mechanomyographic responses to isometric muscle actions. Eur. J. Appl. Physiol. Occup. Physiol. 1998, 78, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.D.; Cramer, J.T.; Egan, A.D.; Hartman, M.J.; Herda, T.J. Time and frequency domain responses of the mechanomyogram and electromyogram during isometric ramp contractions: A comparison of the short-time fourier and continuous wavelet transforms. J. Electromyogr. Kinesiol. 2008, 18, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, M.; Kouzaki, M.; Yoshihisa, T.; Fukunaga, T. Mechanomyogram from the different heads of the quadriceps muscle during incremental knee extension. Eur. J. Appl. Physiol. Occup. Physiol. 1998, 78, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Katsavelis, D.; Joseph Threlkeld, A. Quantifying thigh muscle co-activation during isometric knee extension contractions: Within-and between-session reliability. J. Electromyogr. Kinesiol. 2014, 24, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.-S.; Lee, T.S.; Kim, H.-J. An application of support vector machines in bankruptcy prediction model. Expert Syst. Appl. 2005, 28, 127–135. [Google Scholar] [CrossRef]
- Owolabi, T.O.; Akande, K.O.; Olatunji, S.O. Support vector machines approach for estimating work function of semiconductors: Addressing the limitation of metallic plasma model. Appl. Phys. Res. 2014, 6, 122–132. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Burges, C.C. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Gupta, S.M. Support vector machines based modelling of concrete strength. World Acad. Sci. Eng. Technol. 2007, 36, 305–311. [Google Scholar]
- Pal, M.; Goel, A. Prediction of the end-depth ratio and discharge in semi-circular and circular shaped channels using support vector machines. Flow Meas. Instrum. 2006, 17, 49–57. [Google Scholar] [CrossRef]
- Lin, S.-W.; Ying, K.-C.; Chen, S.-C.; Lee, Z.-J. Particle swarm optimization for parameter determination and feature selection of support vector machines. Expert Syst.Appl. 2008, 35, 1817–1824. [Google Scholar] [CrossRef]
- Mahmoud, S.A.; Olatunji, S.O. Automatic recognition of off-line handwritten arabic (Indian) numerals using support vector and extreme learning machines. Int. J. Imaging 2009, 2, 34–53. [Google Scholar]
- Akande, K.O.; Owolabi, T.O.; Olatunji, S.O. Investigating the effect of correlation-based feature selection on the performance of support vector machines in reservoir characterization. J. Nat. Gas Sci. Eng. 2015, 22, 515–522. [Google Scholar] [CrossRef]
- Akbani, R.; Kwek, S.; Japkowicz, N. Applying support vector machines to imbalanced datasets. In Machine Learning: Ecml 2004; Boulicaut, J.-F., Esposito, F., Giannotti, F., Pedreschi, D., Eds.; Springer: Berlin, Germany, 2004. [Google Scholar]
- Cherkassky, V.; Ma, Y. Practical selection of svm parameters and noise estimation for svm regression. Neural Netw. 2004, 17, 113–126. [Google Scholar] [CrossRef]
- Rıza Yıldız, A. A novel hybrid immune algorithm for global optimization in design and manufacturing. Robot. Comput.-Integr. Manuf. 2009, 25, 261–270. [Google Scholar] [CrossRef]
- Owolabi, T.O.; Akande, K.O.; Olatunji, S.O. Estimation of surface energies of hexagonal close packed metals using computational intelligence technique. Appl. Soft Comput. 2015, 31, 360–368. [Google Scholar] [CrossRef]
- Olatunji, S.O.; Selamat, A.; Abdul Raheem, A.A. Improved sensitivity based linear learning method for permeability prediction of carbonate reservoir using interval type-2 fuzzy logic system. Appl. Soft Comput. 2014, 14, 144–155. [Google Scholar] [CrossRef]
- Gencoglu, M.T.; Uyar, M. Prediction of flashover voltage of insulators using least squares support vector machines. Expert Syst. Appl. 2009, 36, 10789–10798. [Google Scholar] [CrossRef]
- Tay, F.E.H.; Cao, L. Application of support vector machines in financial time series forecasting. Omega 2001, 29, 309–317. [Google Scholar] [CrossRef]
- Gregory, C.M.; Bickel, C.S. Recruitment patterns in human skeletal muscle during electrical stimulation. Phys. Ther. 2005, 85, 358–364. [Google Scholar] [PubMed]
| C | 879 |
|---|---|
| Hyper-parameter (Lambda) | 2−15 |
| Epsilon () | 0.1205 |
| Kernel option | 54 |
| Kernel | Gaussian (RBF) |
| Stimulation Intensity (mA) | Knee Angle | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 30° | 60° | 90° | |||||||
| PT | RMS | PTP | PT | RMS | PTP | PT | RMS | PTP | |
| 20 | 13.9 (3.7) | 14.7 (9.9) | 23.6 (16.4) | 4.1 (0.7) | 17.4 (18.4) | 21.7 (23.0) | 4.3 (5.7) | 20.4 (20.0) | 22.0 (26.9) |
| 30 | 23.3 (19.7) | 51.9 (22.4) | 55.8 (24.7) | 9.7 (8.5) | 37.4 (21.2) | 38.7 (19.5) | 11.0 (10.2) | 51.3 (30.0) | 50.8 (30.6) |
| 40 | 58.2 (23.6) | 75.3 (29.5) | 73.54 (19.1) | 27.6 (24.2) | 77.7 (36.6) | 65.88 (19.2) | 21.4 (15.0) | 93.4 (45.4) | 84.0 (34.1) |
| 50 | 76.6 (19.3) | 84.2 (15.2) | 85.04 (14.9) | 51.5 (26.2) | 82.6 (27.3) | 72.7 (14.9) | 40.7 (18.5) | 115.7 (39.6) | 101.0 (33.8) |
| 60 | 86.1 (20.2) | 104.9 (22.5) | 94.86 (18.2) | 74.7 (19.2) | 94.9 (30.4) | 85.27 (14.7) | 62.1 (12.3) | 104.3 (29.0) | 101.1 (28.5) |
| 70 | 91.1 (21.5) | 100.2 (6.2) | 98.34 (5.7) | 91.0 (8.2) | 88.1 (9.7) | 90.57 (14.4) | 84.2 (12.3) | 118.5 (22.0) | 113.2 (10.7) |
| 80 | 100.0 (0) | 100.0 (0) | 100.0 (0) | 100.0 (0) | 100.0 (0) | 100.0 (0) | 100.0 (0) | 100.0 (0) | 100.0 (0) |
| Input Parameters | Mean | Max | Median | Stdev | Min |
|---|---|---|---|---|---|
| Participants | |||||
| Weight (kg) | 70.1 | 80 | 69 | 5.9 | 63 |
| Age (years) | 23.4 | 25 | 23.5 | 1.3 | 21 |
| Stimulation intensity (mA) | 50 | 80 | 50 | 20 | 20 |
| Knee angle (°) | 60 | 90 | 60 | 24.5 | 30 |
| Normalized MMG-RMS% | 77.8 | 188.1 | 86.9 | 40.0 | 4 |
| Normalized MMG-PTP% | 75.2 | 163.5 | 81.6 | 34.8 | 4.6 |
| Peak torque | 53.9 | 108.4 | 57.2 | 38 | 0 |
| Performance Measures | Training | Testing |
|---|---|---|
| r | 0.97 | 0.94 |
| R2 | 94% | 89% |
| RMSE | 9.48 | 12.95 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibitoye, M.O.; Hamzaid, N.A.; Abdul Wahab, A.K.; Hasnan, N.; Olatunji, S.O.; Davis, G.M. Estimation of Electrically-Evoked Knee Torque from Mechanomyography Using Support Vector Regression. Sensors 2016, 16, 1115. https://doi.org/10.3390/s16071115
Ibitoye MO, Hamzaid NA, Abdul Wahab AK, Hasnan N, Olatunji SO, Davis GM. Estimation of Electrically-Evoked Knee Torque from Mechanomyography Using Support Vector Regression. Sensors. 2016; 16(7):1115. https://doi.org/10.3390/s16071115
Chicago/Turabian StyleIbitoye, Morufu Olusola, Nur Azah Hamzaid, Ahmad Khairi Abdul Wahab, Nazirah Hasnan, Sunday Olusanya Olatunji, and Glen M. Davis. 2016. "Estimation of Electrically-Evoked Knee Torque from Mechanomyography Using Support Vector Regression" Sensors 16, no. 7: 1115. https://doi.org/10.3390/s16071115
APA StyleIbitoye, M. O., Hamzaid, N. A., Abdul Wahab, A. K., Hasnan, N., Olatunji, S. O., & Davis, G. M. (2016). Estimation of Electrically-Evoked Knee Torque from Mechanomyography Using Support Vector Regression. Sensors, 16(7), 1115. https://doi.org/10.3390/s16071115






