Arrhythmia Classification Based on Multi-Domain Feature Extraction for an ECG Recognition System
Abstract
:1. Introduction
2. Materials and Methods
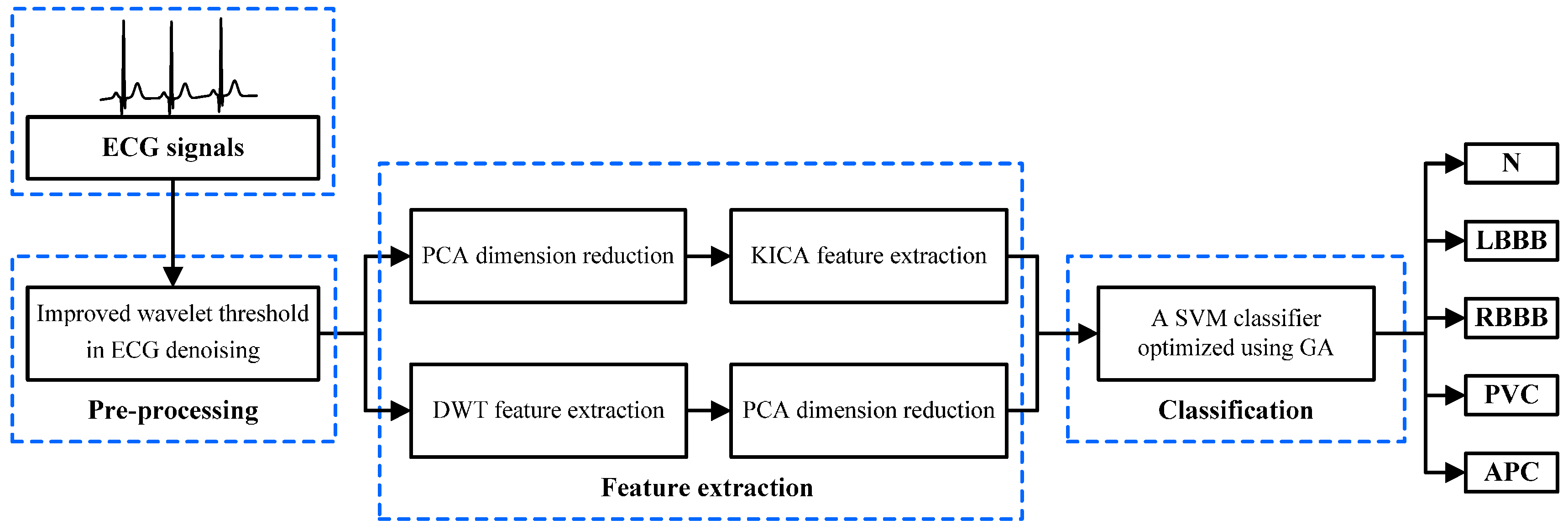
2.1. The Proposed System Based on the Multi-Domain Feature Extraction
- 1
- The ECG beats are pre-processed to eliminate disturbance and noise by denoising with an improved wavelet threshold method.
- 2
- The original ECG data are optimized through PCA. KICA is applied to reduce data dimensions and obtain the nonlinear features of ECG beats. The DWT method is employed to extract frequency domain features. The maximum, minimum, mean and standard deviation values of each sampling signal wavelet coefficient are calculated. Additionally, LDA is used to optimise the frequency domain features.
- 3
- The multi-domain features are composed of nonlinear and frequency domain features, which are used as input features to train and test the SVM classifier model. GA is employed to optimise the SVM parameters and improve the classifier’s performance. Finally, the five types of ECG beats derived from the MIT-BIH arrhythmia database are classified with the optimised SVM classifier.
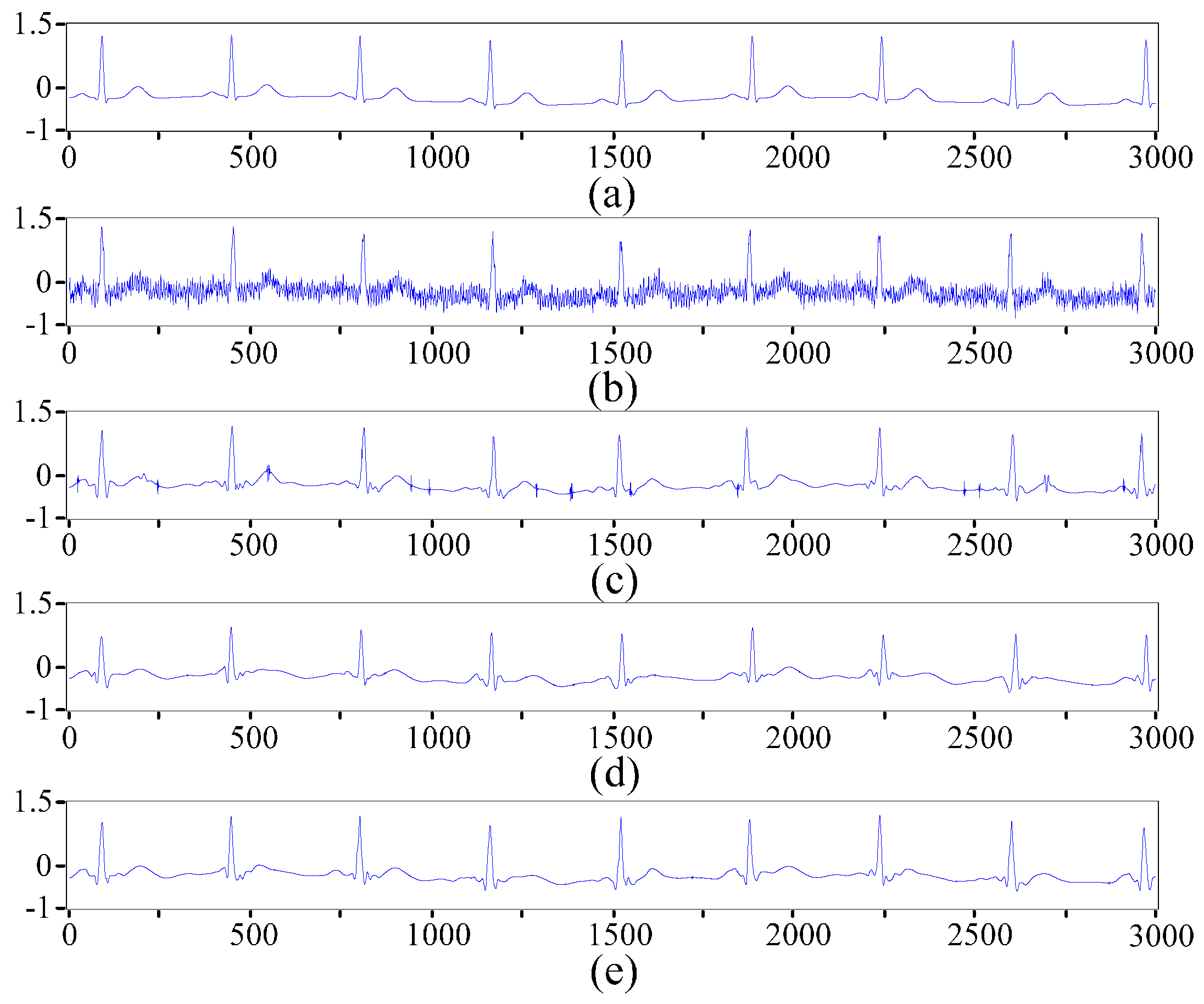
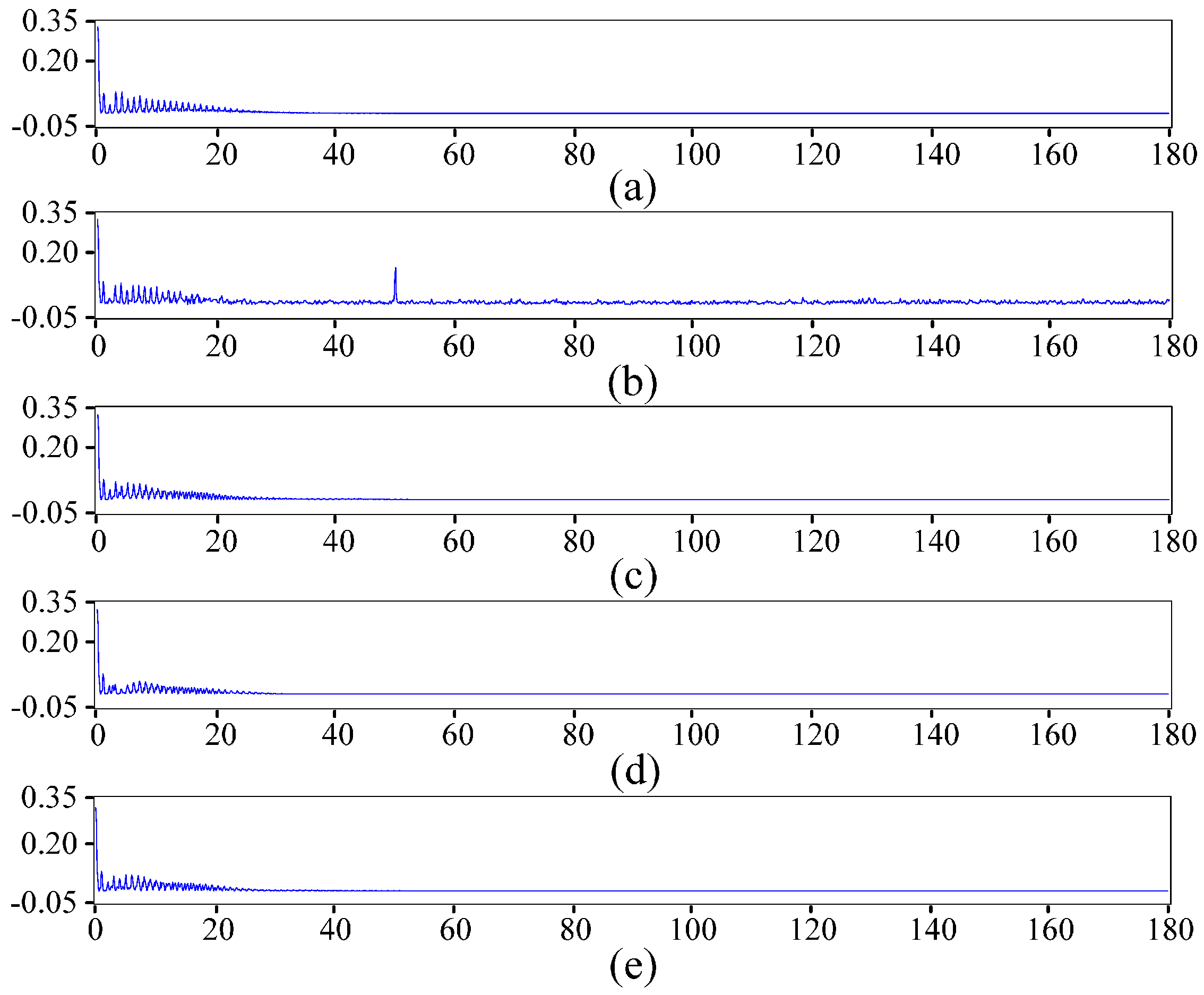
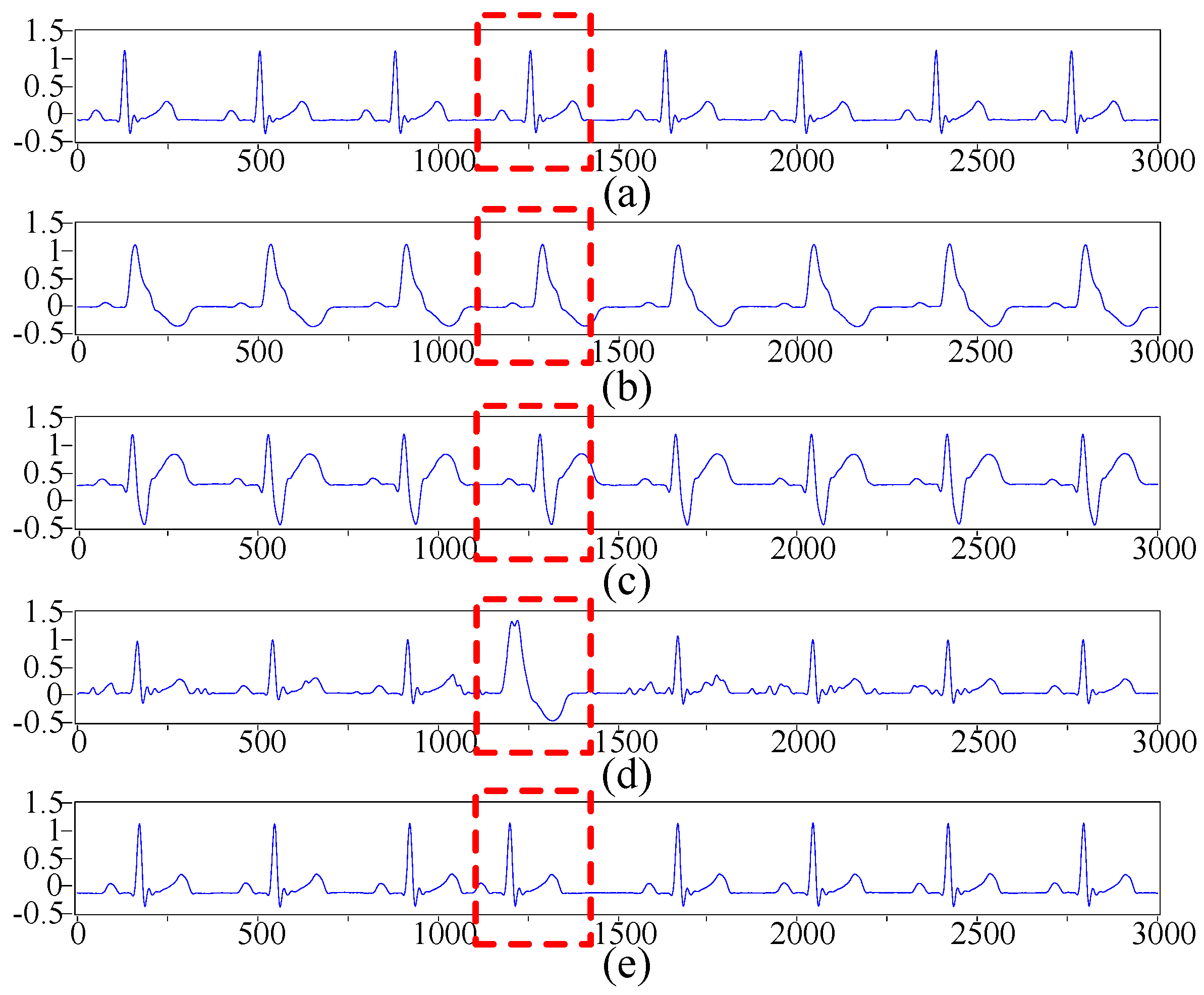
2.2. ECG Pre-Processing Based on the Improved Wavelet Threshold Method
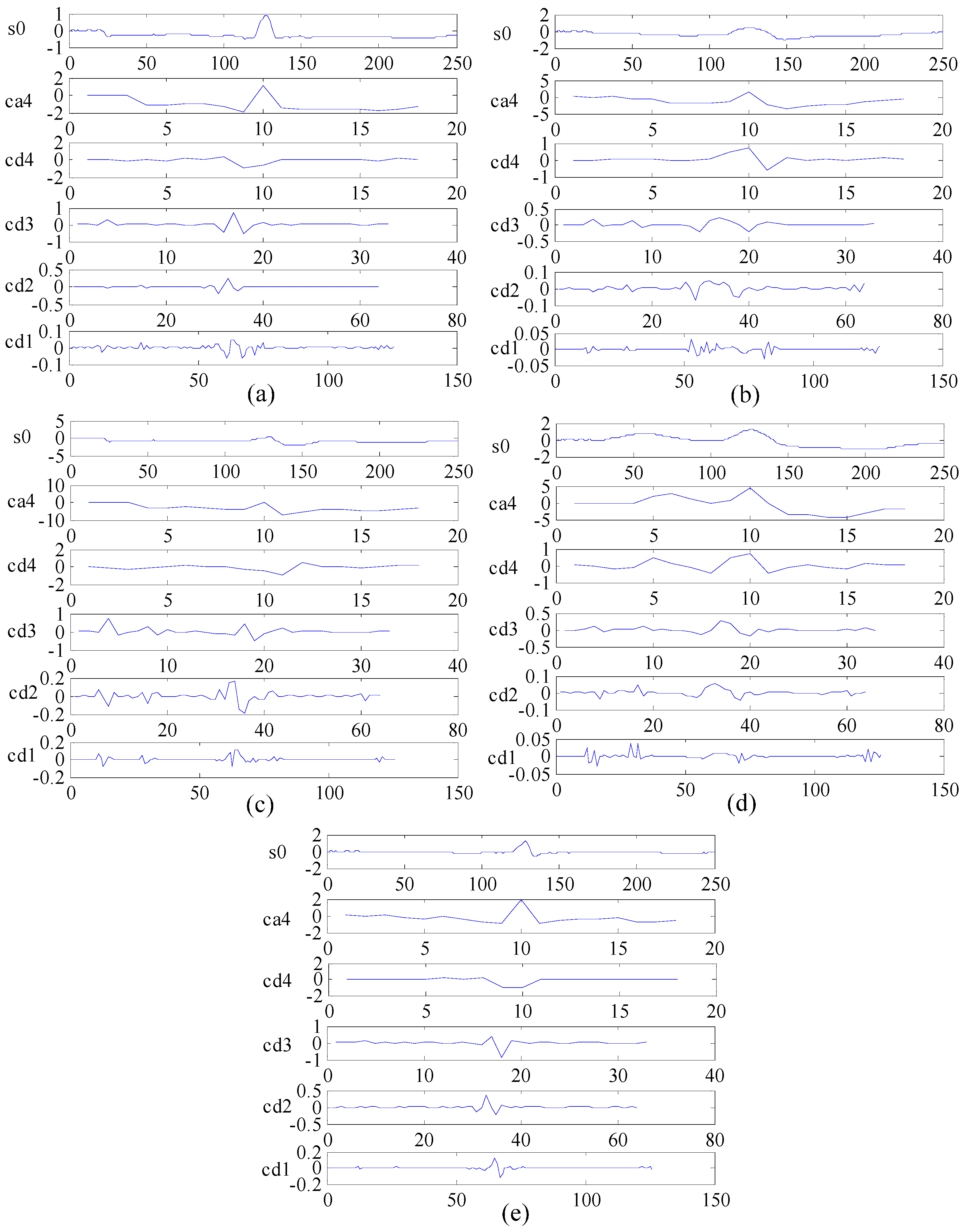
2.3. Multi-Domain Feature Extraction
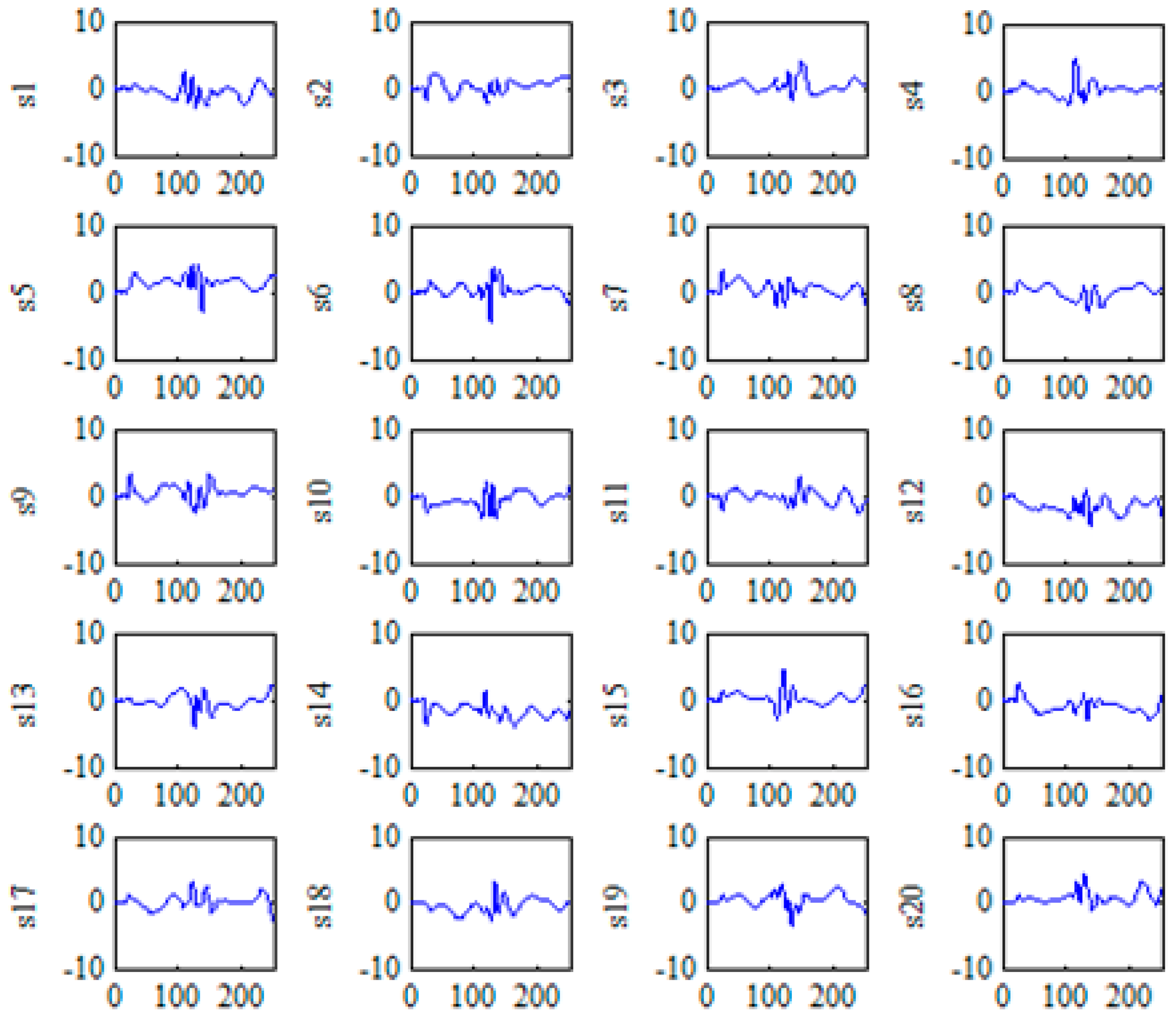
2.3.1. KICA for the Nonlinear Feature Extraction
- Enter ECG data and determine the number of sources p.
- Initialize the decomposition matrix W.
- Evaluate the source signals, si = Wxi, and compute the centralized Gram matrix, K1, …, Kp. Here, Ki = k(xi,x) (I = 1, 2, …, p).
- Compute the minimum eigenvalue, λM (K1, …, Kp), of the generalized eigenvector equation, Kα = λDα.
- Compute the target function:
- Minimize the target function C(W) and output W.
2.3.2. DWT for Frequency Domain Feature Extraction
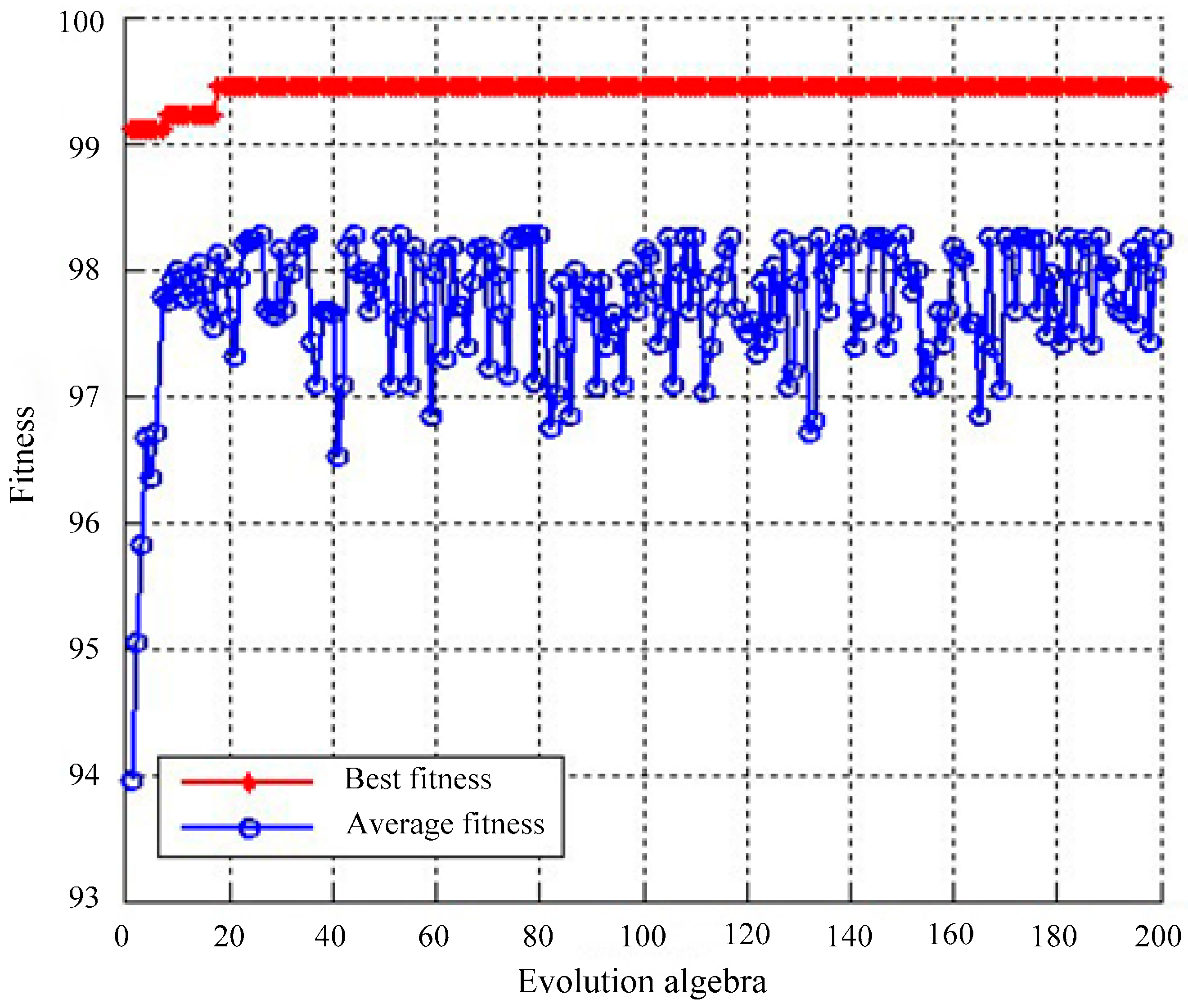
2.4. Classification Based on SVM Optimized by GA
- Generate an initial population with the binary code.
- Compute the fitness function and the training set for CV to determine the fitness function value.
- Filter a new population from the old population on the basis of individual fitness, which is determined by the evaluation function.
- Employ several genetic operators of mutation and crossover to generate new solutions.
- Calculate the fitness function of the newly generated individuals.
- Repeat steps 3–5 until the maximum iteration is reached and then find the optimal parameters.
3. Experimental Design
4. Results and Discussion
4.1. Results of the Proposed ECG Recognition System
4.1.1. Multi-Domain Feature Extraction Using KICA Combined with DWT
4.1.2. Classification Using SVM Optimized by GA
4.2. Experimental Results from the ECG Acquisition Platform
4.3. Discussion and Comparisons
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Edla, S.; Kovvali, N.; Papandreou-Suppappola, A. Electrocardiogram signal modeling with adaptive parameter estimation using sequential bayesian methods. IEEE Trans. Signal Process. 2014, 62, 2667–2680. [Google Scholar] [CrossRef]
- Wang, J.; She, M.; Nahavandi, S.; Kouzani, A. Human identification from ECG signals via sparse representation of local segments. IEEE Signal Process. Lett. 2013, 20, 937–940. [Google Scholar] [CrossRef]
- Ubeyli, E.D. Combining recurrent neural networks with eigenvector methods for classification of ECG beats. Digit. Signal Process. 2009, 19, 320–329. [Google Scholar] [CrossRef]
- Jain, S.; Bajaj, V.; Kumar, A. Efficient algorithm for classification of electrocardiogram beats based on artificial bee colony-based least-squares support vector machines classifier. Electron. Lett. 2016, 52, 1198–1200. [Google Scholar] [CrossRef]
- Zadeh, A.E.; Khazaee, A. High efficient system for automatic classification of the electrocardiogram beats. Ann. Biomed. Eng. 2011, 39, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Plonsey, R.; Barr, R.C. Bioelectricity: A Quantitative Approach; Springer: New York, NY, USA, 2000; pp. 522–526. [Google Scholar]
- Perlman, O.; Katz, A.; Weissman, N.; Amit, G. Atrial electrical activity detection using linear combination of 12-lead ECG signals. IEEE Trans. Biomed. Eng. 2014, 61, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Afonso, V.X.; Tompkins, W.J.; Nguyen, T.Q.; Luo, S. ECG beat detection using filter banks. IEEE Trans. Biomed. Eng. 1999, 46, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Torres, A.; Soria, C.; Santos, J.L. A method for removing noise from continuous brain signal recordings. Comput. Electr. Eng. 2013, 39, 1561–1570. [Google Scholar] [CrossRef]
- Mateo, J.; Sanchez-Morla, E.M.; Santos, J.L. A new method for removal of powerline interference in ECG and EEG recordings. Comput. Electr. Eng. 2015, 45, 235–248. [Google Scholar] [CrossRef]
- Liu, S.; Jilai, L.U.; Hao, L.; Guangshu, H.U. Detection of QRS complex using mathematical morphology and wavelet transform. J. Tsinghua Univ. 2004, 44, 852–855. [Google Scholar]
- Vizireanu, D.N. Morphological shape decomposition interframe interpolation method. J. Electron. Imaging. 2008, 17, 1–5. [Google Scholar] [CrossRef]
- Szilagyi, S.M.; Benyo, Z.; Szilagyi, L.; David, L. Adaptive wavelet-transform-based ECG waveforms detection. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Cancún, Mexico, 17–21 September 2003; pp. 2412–2415.
- Alfaouri, M.; Daqrouq, K. ECG signal denoising by wavelet transform thresholding. Am. J. Appl. Sci. 2008, 5, 276–281. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, L.; Chen, D.; Luo, Y. A human ECG identification system based on ensemble empirical mode decomposition. Sensors 2013, 13, 6832–6864. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Q.; Wang, X.F. Detection of electrocardiogram characteristic points using lifting wavelet transform and Hilbert transform. Trans. Inst. Meas. Control. 2013, 35, 574–582. [Google Scholar] [CrossRef]
- Gao, J.; Sultan, H.; Hu, J.; Tung, W.W. Denoising nonlinear time series by adaptive filtering and wavelet shrinkage: A comparison. IEEE Signal Process. Lett. 2010, 17, 237–240. [Google Scholar]
- Li, H.Q.; Wang, X.F.; Chen, L.; Li, E.B. Denoising and R-Peak detection of electrocardiogram signal based on EMD and improved approximate envelope. Circ. Syst. Signal Process. 2014, 33, 1261–1276. [Google Scholar] [CrossRef]
- Ganeshkumar, R.; Yskumaraswamy, D. Investigating cardiac arrhythmia in ECG using random forest classification. Int. J. Comput. Appl. 2012, 37, 31–34. [Google Scholar] [CrossRef]
- Fei, S.W. Diagnostic study on arrhythmia cordis based on particle swarm optimization-based support vector machine. Expert Syst. Appl. 2010, 37, 6748–6752. [Google Scholar] [CrossRef]
- Liu, S.H.; Cheng, D.C.; Lin, C.M. Arrhythmia identification with two-lead electrocardiograms using artificial neural networks and support vector machines for a portable ECG monitor system. Sensors 2013, 13, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Rai, H.M.; Trivedi, A.; Shukla, S. ECG signal processing for abnormalities detection using multi-resolution wavelet transform and artificial neural network classifier. Measurement 2013, 46, 3238–3246. [Google Scholar] [CrossRef]
- Daamouche, A.; Hamami, L.; Alajlan, N.; Melgani, F. A wavelet optimization approach for ECG signal classification. Biomed. Signal Process. Control 2012, 7, 342–349. [Google Scholar] [CrossRef]
- Lin, C.C.; Yang, C.M. Heartbeat classification using normalized RR intervals and morphological features. Math. Probl. Eng. 2014, 12, 1–11. [Google Scholar] [CrossRef]
- Dutta, S.; Chatterjee, A.; Munshi, S. Correlation technique and least square support vector machine combine for frequency domain based ECG beat classification. Med. Eng. Phys. 2010, 32, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Kasturiwale, H.P.; Ingole, P.V. Component extraction of complex biomedical signals and performance analysis. Int. J. Comput. Sci. Inf. Technol. 2012, 3, 3544–3547. [Google Scholar]
- Martis, R.J.; Acharya, U.R.; Mandana, K.M. Cardiac decision making using higher order spectra. Biomed. Signal Process. 2013, 8, 193–203. [Google Scholar] [CrossRef]
- Valenza, G.; Citi, L.; Lanata, A.; Scilingo, E.P.; Barbieri, R. Revealing real-time emotional responses: A personalized assessment based on heartbeat dynamics. Sci. Rep. 2014, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Saini, I.; Singh, D.; Khosla, A. Electrocardiogram beat classification using empirical mode decomposition and multi-class directed acyclic graph support vector machine. Comput. Electr. Eng. 2014, 40, 1774–1787. [Google Scholar] [CrossRef]
- Kamath, C. ECG beat classification using features extracted from Teager energy functions in time and frequency domains. IET Signal Process. 2011, 5, 575–581. [Google Scholar] [CrossRef]
- Bach, F.R.; Jordan, M.I. Kernel independent component analysis. J. Mach. Learn. Res. 2003, 3, 1–48. [Google Scholar]
- Belhumeur, P.; Hespanha, J.; Kriegman, D. Eigenfaces vs. fisherfaces: Recognition using class specific linear projection. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 19, 711–720. [Google Scholar] [CrossRef]
- Goldberg, D.E. Genetic Algorithms in Search, Optimization, and Machine Learning; Addison Wesley: Boston, MA, USA, 1989; pp. 2104–2116. [Google Scholar]
- Goldberger, A.L. Clinical Electrocardiography: A Simplified Approach; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Donoho, D.L. De-noising by soft-thresholding. IEEE Trans. Inform. Theory 1995, 41, 613–627. [Google Scholar] [CrossRef]
- Mallat, S.G. A theory for multiresolution signal decomposition: The wavelet representation. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 11, 674–693. [Google Scholar] [CrossRef]
- Jovic, A.; Bogunovic, N. Electrocardiogram analysis using a combination of statistical, geometric, and nonlinear heart rate variability features. Artif. Intell. Med. 2011, 51, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; Wiley: NewYork, NY, USA, 2001; pp. 119–131. [Google Scholar]
- Ubeyli, E.D. ECG beats classification using multiclass support vector machines with error correcting output codes. Digit. Signal Process. 2007, 17, 675–684. [Google Scholar] [CrossRef]
- Vapnik, V. Statistical Learning Theory; Wiley: NewYork, NY, USA, 1998. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Gamarra, A.; Quintero, M. Using genetic algorithm feature selection in neural classification systems for image pattern recognition. Ing. Investig. 2013, 33, 52–58. [Google Scholar]
- Huang, C.J. Using genetic algorithm optimization SVM to construction of investment model. Int. J. Digit. Content Technol. Appl. 2011, 5, 123–132. [Google Scholar]
- Zadeh, A.E.; Khazaee, A.; Ranaee, V. Classification of the electrocardiogram signals using supervised classifiers and efficient features. Comput. Meth. Program. Biomed. 2010, 99, 179–194. [Google Scholar] [CrossRef] [PubMed]


| Methods | Performance Indicators | ECG Signal Records | ||||
|---|---|---|---|---|---|---|
| 103 | 102 | 118 | 232 | 231 | ||
| Hard threshold | signal-noise ratio (SNR) | 21.2769 | 20.4320 | 21.8957 | 18.1098 | 22.6279 |
| root mean square error (RMSE) | 0.0255 | 0.0252 | 0.0257 | 0.0319 | 0.0221 | |
| Soft threshold | SNR | 21.7454 | 20.2268 | 22.1032 | 18.0057 | 22.5574 |
| RMSE | 0.0241 | 0.0258 | 0.0251 | 0.0328 | 0.0223 | |
| Improved threshold | SNR | 24.0626 | 23.4869 | 24.0128 | 20.8680 | 25.2851 |
| RMSE | 0.0185 | 0.0178 | 0.0179 | 0.0236 | 0.0163 | |
| Type | N | LBBB | RBBB | PVC | APC |
|---|---|---|---|---|---|
| normal beat (N) | 200 | 0 | 0 | 0 | 0 |
| left bundle branch block beat (LBBB) | 0 | 198 | 0 | 2 | 0 |
| right bundle branch block beat (RBBB) | 0 | 0 | 200 | 0 | 0 |
| premature ventricular contraction (PVC) | 0 | 5 | 0 | 195 | 0 |
| atrial premature beat (APC) | 3 | 0 | 0 | 1 | 96 |
| Type | Sensitivity (Se) | Specificity (Sp) | Positive Predictability (Pp) |
|---|---|---|---|
| normal beat (N) | 100% | 99.57% | 98.52% |
| LBBB | 99% | 99.29% | 97.54% |
| RBBB | 100% | 100% | 100% |
| PVC | 97.50% | 99.57% | 98.48% |
| APC | 96% | 100% | 100% |
| Average | 98.50% | 99.69% | 98.91% |
| Accuracy(Acc) | 98.80% |
| Type | N | LBBB | RBBB | PVC | APC |
|---|---|---|---|---|---|
| N | 195 | 2 | 0 | 1 | 2 |
| LBBB | 4 | 194 | 0 | 2 | 0 |
| RBBB | 0 | 0 | 199 | 1 | 0 |
| PVC | 1 | 8 | 2 | 189 | 0 |
| APC | 1 | 0 | 0 | 0 | 99 |
| Type | Se | Sp | Pp |
|---|---|---|---|
| N | 97.5% | 99.14% | 97.01% |
| LBBB | 97% | 98.57% | 95.10% |
| RBBB | 99.50% | 99.71% | 99% |
| PVC | 94.50% | 99.43% | 97.93% |
| APC | 99% | 99.75% | 98.02% |
| Average | 97.50% | 99.32% | 97.41% |
| Acc | 97.30% |
| Methods | Classifier | Classes | Accuracy | Reference |
|---|---|---|---|---|
| Principal components of bispectrum features | least squares support vector machine (LSSVM) | 5 | 93.48% | Martis et al. |
| Teager energy function features | neural network (NN) | 5 | 95% | Kamath |
| Morphological and time features | support vector machine (SVM) | 3 | 97.14% | Zadeh et al. |
| Time intervals | SVM | 5 | 95.65% | Fei |
| R-R intervals | self-constructing neural fuzzy interference network (SoNFIN) | 5 | 96.4% | Liu |
| The multi-domain features | a library for SVM (LIBSVM) | 5 | 98.8% | Proposed method |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Yuan, D.; Wang, Y.; Cui, D.; Cao, L. Arrhythmia Classification Based on Multi-Domain Feature Extraction for an ECG Recognition System. Sensors 2016, 16, 1744. https://doi.org/10.3390/s16101744
Li H, Yuan D, Wang Y, Cui D, Cao L. Arrhythmia Classification Based on Multi-Domain Feature Extraction for an ECG Recognition System. Sensors. 2016; 16(10):1744. https://doi.org/10.3390/s16101744
Chicago/Turabian StyleLi, Hongqiang, Danyang Yuan, Youxi Wang, Dianyin Cui, and Lu Cao. 2016. "Arrhythmia Classification Based on Multi-Domain Feature Extraction for an ECG Recognition System" Sensors 16, no. 10: 1744. https://doi.org/10.3390/s16101744
APA StyleLi, H., Yuan, D., Wang, Y., Cui, D., & Cao, L. (2016). Arrhythmia Classification Based on Multi-Domain Feature Extraction for an ECG Recognition System. Sensors, 16(10), 1744. https://doi.org/10.3390/s16101744







