A Sensitive and Selective Label-Free Electrochemical DNA Biosensor for the Detection of Specific Dengue Virus Serotype 3 Sequences
Abstract
:1. Introduction
2. Experimental Section
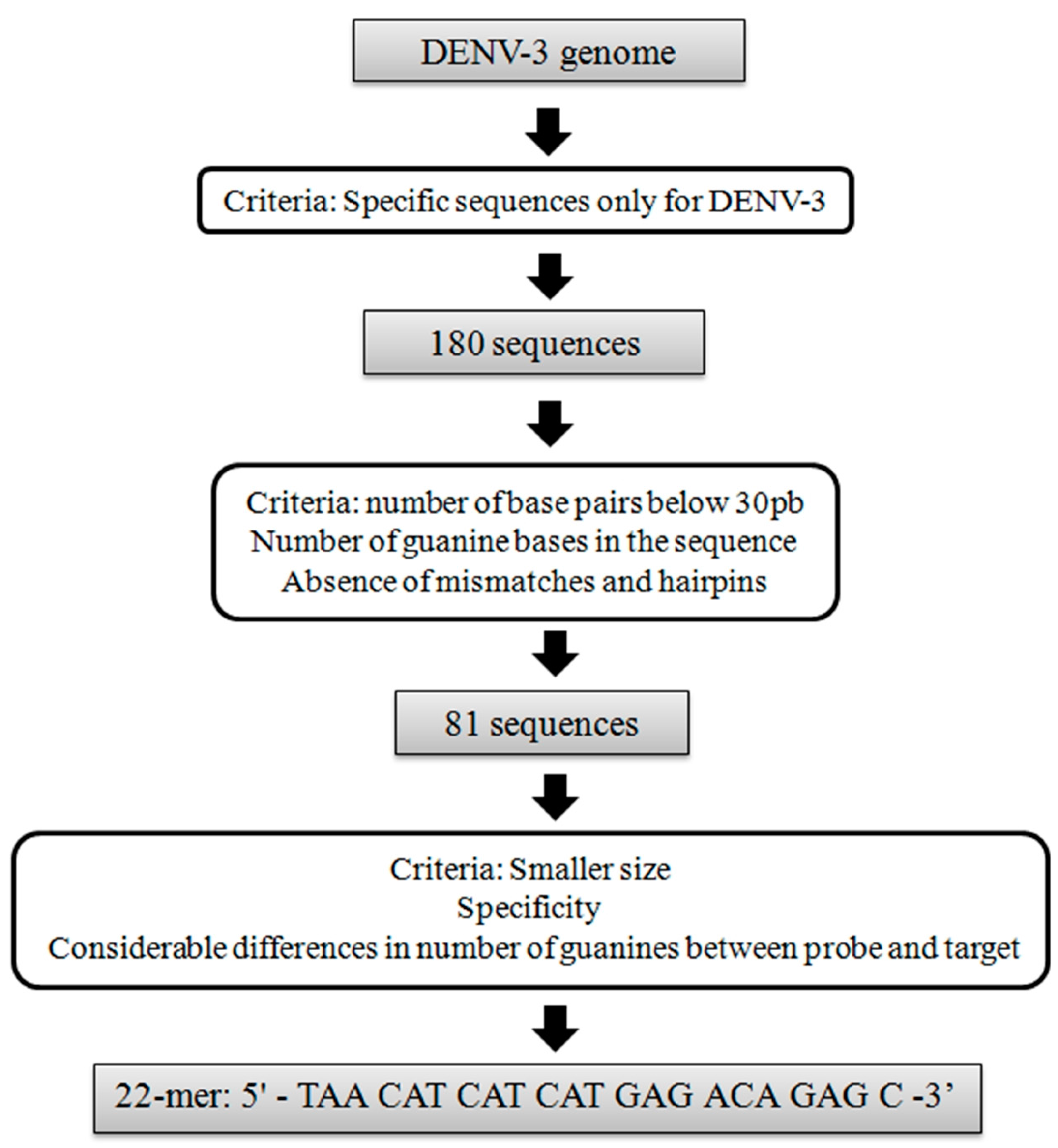
2.1. Design of a Specific DENV-3 DNA Probe
2.2. Reagents and Materials
- DENV-3 probe: 5′-TAA CAT CAT CAT GAG ACA GAG C-3′
- DENV-3 target: 5′-GCT CTG TCT CAT GAT GAT GTT A-3′
- Non-complementary sequence: 5′-TCT CTT GTT TAA GAC AAC AGA G-3′
2.3. Apparatus
2.4. Procedure
2.4.1. Preparation of Electrodes and Pre-Treatment of PGE
2.4.2. DNA Probe Immobilization onto PGE Surface
2.4.3. DNA Hybridization with Complementary and Non-Complementary Sequences
2.4.4. Detection of Complementary and Non-Complementary Sequences in Human Serum
2.5. Electrochemical Analysis
2.6. Statistical Data Analysis
3. Results and Discussion
3.1. Bioinformatics Analysis of DENV-3 DNA Probes
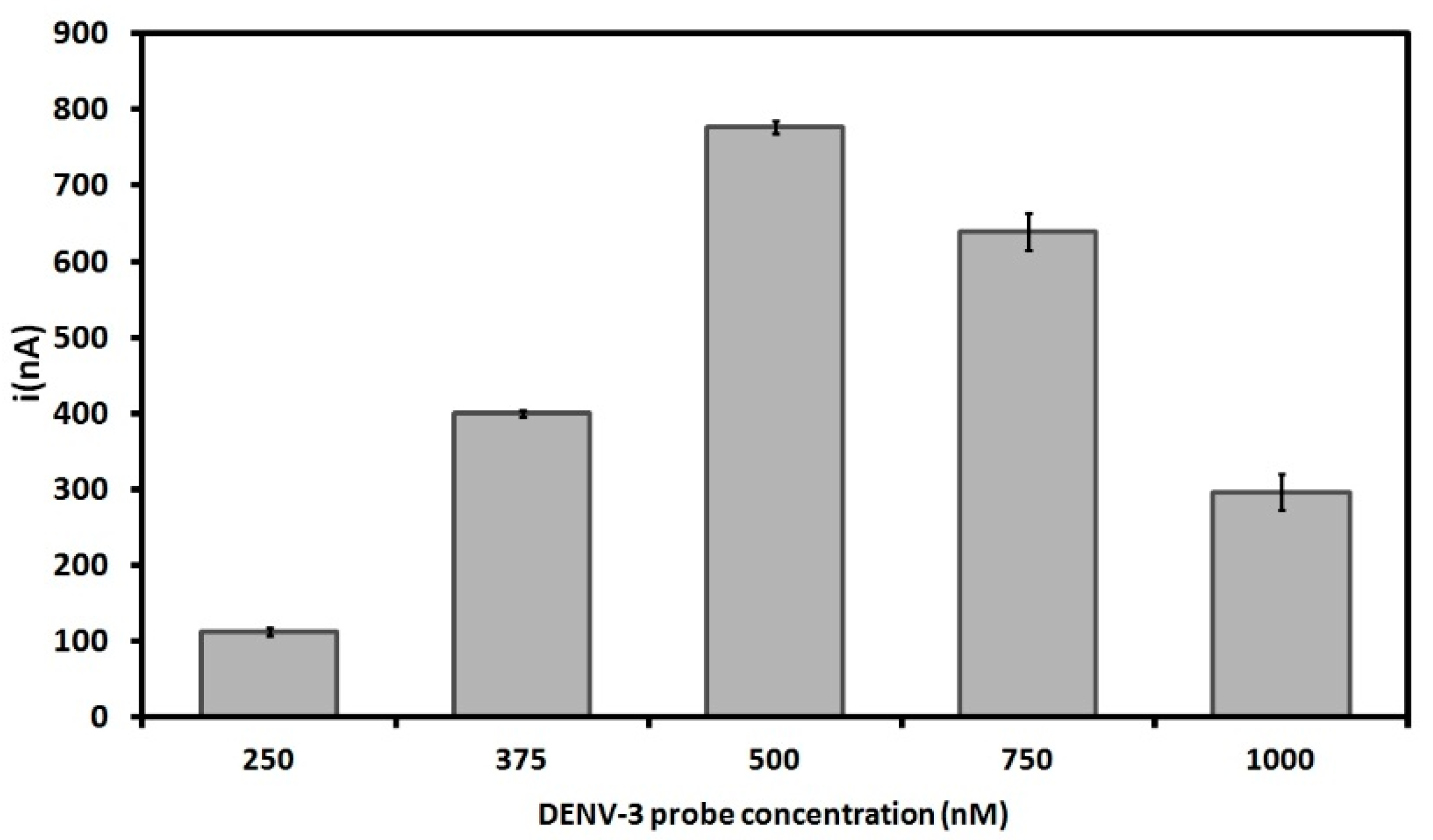
3.2. Effect of DENV-3 Probes Concentration on Immobilization on the PGE
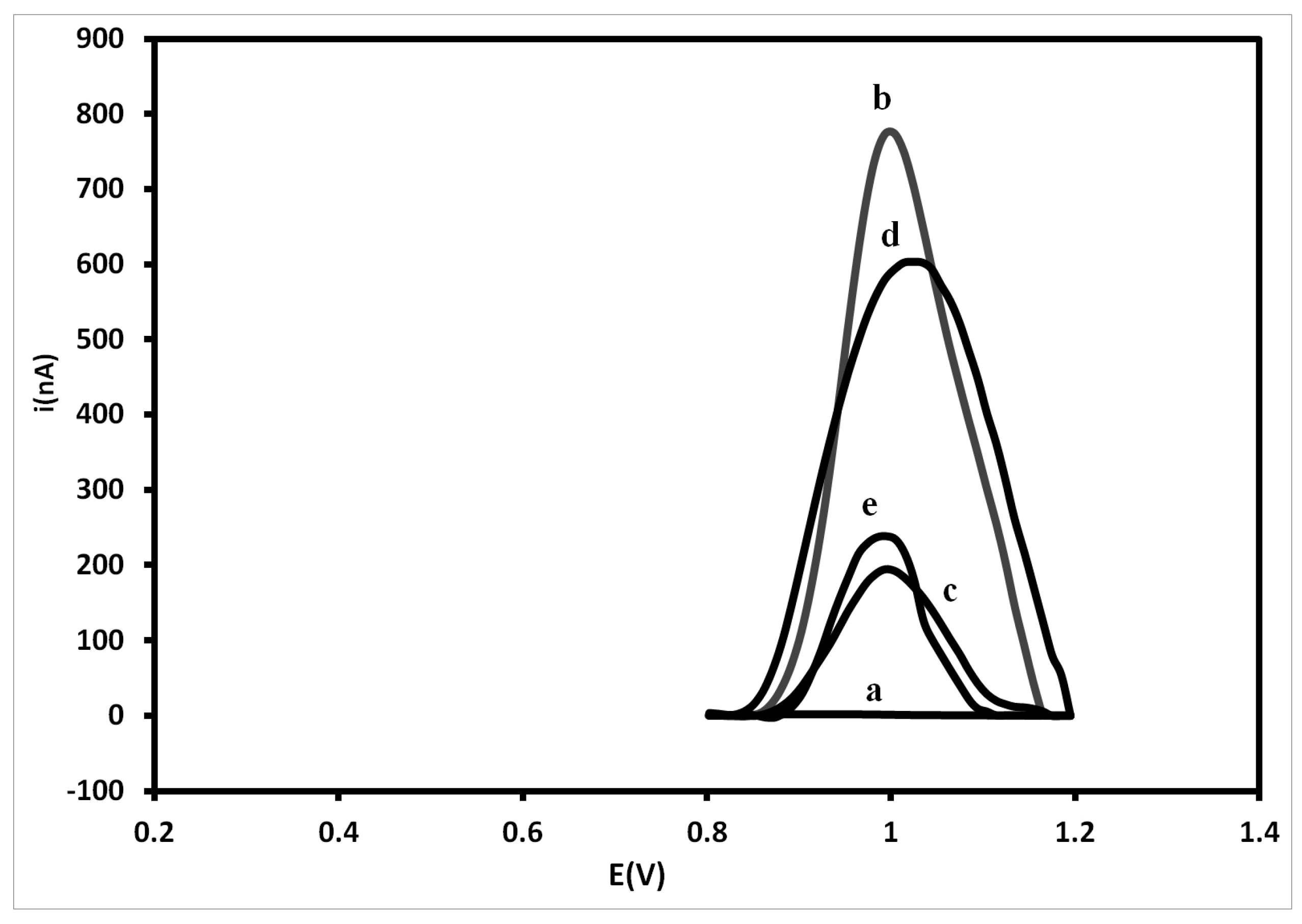
3.3. Electrochemical Analysis of Hybridization Assays


| Nucleic Acid Biosensor | Electrode | Electrochemical Method | Linear Range of Hybridization | Detection Limit | Reference |
|---|---|---|---|---|---|
| Single-walled carbon nanotubes-polymer modified graphite electrodes for DNA hybridization | PGE a | DPV d | 50–200μg/mL | 5.14 μM | [61] |
| Hybridization biosensor for detection of hepatitis B virus | GCE b | DPV | 0.36–1.32 μM | 19.4 nM | [62] |
| Brilliant cresyl blue as electroactive indicator in electrochemical DNA oligonucleotide sensors | CPE c | DPV | 10 nM–5μM | 9 nM | [63] |
| Label-free DNA detection based on zero current potentiometry | PGE | LSV e | 10 nM–1μM | 6.9 nM | [64] |
| DNA biosensor detection of DENV-3 sequences onto PGE surfaces | PGE | DPV | 10–100 nM | 3.09 nM | This work |
3.4. Selectivity Study
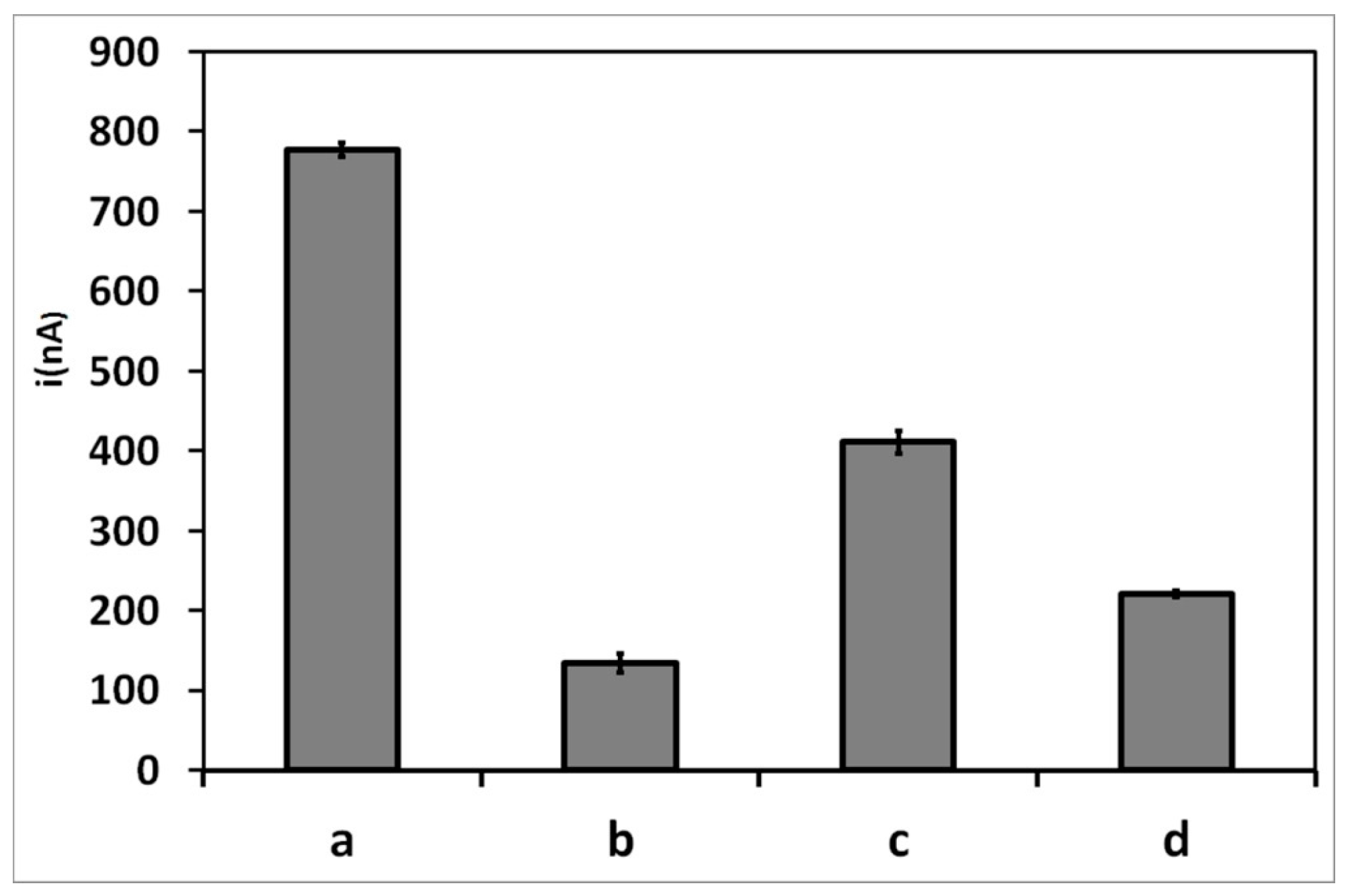
3.5. Electrochemical Measurement of Target Hybridization in Human Serum Solutions
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO. Dengue—Guidelines for Diagnosis, Treatment, Prevention and Control; WHO Press: Geneva, Switzerland, 2009; p. 147. [Google Scholar]
- Sariol, C.A.; White, L.J. Utility, limitations, and future of non-human primates for dengue research and vaccine development. Front. Immunol. 2014, 5, 452. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; Myers, M.F.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar]
- Vasilakis, N.; Cardosa, J.; Hanley, K.A.; Holmes, E.C.; Weaver, S.C. Fever from the forest: Prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 2011, 9, 532–541. [Google Scholar] [CrossRef]
- Chien, L.C.; Yu, H.L. Impact of meteorological factors on the spatiotemporal patterns of dengue fever incidence. Environ. Int. 2014, 73, 46–56. [Google Scholar] [CrossRef]
- Cedillo-Barrón, L.; García-Cordero, J.; Bustos-Arriaga, J.; León-Juárez, M.; Gutiérrez-Castañeda, B. Antibody response to dengue virus. Microbes Infect. 2014, 16, 711–720. [Google Scholar] [CrossRef]
- Rovida, F.; Percivalle, E.; Campanini, G.; Piralla, A.; Novati, S.; Muscatello, A.; Baldanti, F. Viremic Dengue virus infections in travellers: Potential for local outbreak in Northern Italy Dengue-1 Dengue-4. J. Clin. Virol. 2011, 50, 76–79. [Google Scholar] [CrossRef]
- Ashley, E.A. Trends in Anaesthesia and Critical Care Dengue fever. Trends Anaesth. Crit. Care 2011, 1, 39–41. [Google Scholar] [CrossRef]
- Qing, X.; Sun, N.; Yeh, J.; Yue, C.; Cai, J. Dengue fever and bone marrow myelofibrosis. Exp. Mol. Pathol. 2014, 97, 208–210. [Google Scholar] [CrossRef]
- Halstead, S.B. Dengue. Lancet 2007, 370, 1644–1652. [Google Scholar] [CrossRef]
- Arora, P.; Sindhu, A.; Dilbaghi, N.; Chaudhury, A. Biosensors as innovative tools for the detection of food borne pathogens. Biosens. Bioelectron. 2011, 28, 1–12. [Google Scholar] [CrossRef]
- Chakravarti, A.; Arora, R.; Luxemburger, C. Fifty years of dengue in India. Trans. R. Soc. Trop. Med. Hyg. 2012, 106, 273–282. [Google Scholar] [CrossRef]
- Verma, R.; Sahu, R.; Holla, V. Neurological manifestations of dengue infection: A review. J. Neurol. Sci. 2014, 346, 26–34. [Google Scholar] [CrossRef]
- Pechansky, F.; Duarte, P.D.C.A.V.; de Boni, R.; Leukefeld, C.G.; von Diemen, L.; Bumaguin, D.B.; Kreische, F.; Hilgert, J.B.; Bozzetti, M.C.; Fuchs, D.F.P. Predictors of positive Blood Alcohol Concentration (BAC) in a sample of Brazilian drivers. Rev. Bras. Psiquiatr. 2012, 34, 277–285. [Google Scholar] [CrossRef]
- Ferraz, F.O.; Bomfim, M.R.Q.; Totola, A.H.; Ávila, T.V.; Cisalpino, D.; Pessanha, J.E.M.; da Glória de Souza, D.; Teixeira Júnior, A.L.; Nogueira, M.L.; Bruna-Romero, O.; et al. Evaluation of laboratory tests for dengue diagnosis in clinical specimens from consecutive patients with suspected dengue in Belo Horizonte, Brazil. J. Clin. Virol. 2013, 58, 41–46. [Google Scholar]
- Peeling, R.W.; Artsob, H.; Pelegrino, J.L.; Buchy, P.; Cardosa, M.J.; Devi, S.; Enria, D.A.; Farrar, J.; Gubler, D.J.; Guzman, M.G.; et al. Evaluation of diagnostic tests: Dengue. Nat. Rev. Microbiol. 2010, 8, S30–S37. [Google Scholar]
- Wattal, C.; Goel, N. Infectious Disease Emergencies in Returning Travelers—Special Reference to Malaria, Dengue Fever and Chikungunya. Med. Clin. North Am. 2012, 96, 1225–1255. [Google Scholar] [CrossRef]
- Korhonen, E.M.; Huhtamo, E.; Virtala, A.M.K.; Kantele, A.; Vapalahti, O. Approach to non-invasive sampling in dengue diagnostics: Exploring virus and NS1 antigen detection in saliva and urine of travelers with dengue. J. Clin. Virol. 2014, 61, 353–358. [Google Scholar] [CrossRef]
- Shenoy, B.; Menon, A.; Biradar, S. Science Direct Diagnostic utility of dengue NS1 antigen. Pediatr. Infect. Dis. 2014, 6, 110–113. [Google Scholar] [CrossRef]
- Hapugoda, M.D.; de Silva, N.R.; Khan, B.; Damsiri Dayanath, M.Y.; Gunesena, S.; Prithimala, L.D.; Abeyewickreme, W. A comparative retrospective study of RT-PCR-based liquid hybridization assay for early, definitive diagnosis of dengue. Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 279–282. [Google Scholar] [CrossRef]
- Back, A.T.; Lundkvist, A. Dengue viruses—An overview. Infect. Ecol. Epidemiol. 2013, 1, 1–21. [Google Scholar] [CrossRef]
- Siddiquee, S.; Rovina, K.; Yusof, N.A.; Rodrigues, K.F. Nanoparticle-enhanced electrochemical biosensor with DNA immobilization and hybridization of Trichoderma harzianum gene. Sens. Bio-Sens. Res. 2014, 2, 16–22. [Google Scholar] [CrossRef]
- Teles, F.S.R.R. Biosensors and rapid diagnostic tests on the frontier between analytical and clinical chemistry for biomolecular diagnosis of dengue disease: A review. Anal. Chim. Acta 2011, 687, 28–42. [Google Scholar] [CrossRef]
- Lucarelli, F.; Tombelli, S.; Minunni, M.; Marrazza, G.; Mascini, M. Electrochemical and piezoelectric DNA biosensors for hybridisation detection. Anal. Chim. Acta 2008, 609, 139–159. [Google Scholar] [CrossRef]
- Souada, M.; Piro, B.; Reisberg, S.; Anquetin, G.; Noël, V.; Pham, M.C. Label-free electrochemical detection of prostate-specific antigen based on nucleic acid aptamer. Biosens. Bioelectron. 2014, 68C, 49–54. [Google Scholar] [CrossRef]
- Tosar, J.P.; Keel, K.; Laíz, J. Two independent label-free detection methods in one electrochemical DNA sensor. Biosens. Bioelectron. 2009, 24, 3036–3042. [Google Scholar] [CrossRef]
- Sadik, O.A.; Aluoch, A.O.; Zhou, A. Status of biomolecular recognition using electrochemical techniques. Biosens. Bioelectron. 2009, 24, 2749–2765. [Google Scholar]
- Conde, J.; Edelman, E.R.; Artzi, N. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: The jack-of-all-trades in cancer nanotheranostics? Adv. Drug Deliv. Rev. 2015, 81C, 169–183. [Google Scholar] [CrossRef]
- Souza, E.; Nascimento, G.; Santana, N.; Ferreira, D.; Lima, M.; Natividade, E.; Martins, D.; Lima-Filho, J. Label-free electrochemical detection of the specific oligonucleotide sequence of dengue virus type 1 on pencil graphite electrodes. Sensors 2011, 11, 5616–5629. [Google Scholar] [CrossRef]
- Lourenço-de-Oliveira, R.; Honório, N.A.; Castro, M.G.; Schatzmayr, H.G.; Miagostovich, M.P.; Alves, J.C.R.; Silva, W.C.; Leite, P.J.; Nogueira, R. Dengue Virus Type 3 Isolation from Aedes aegypti in the Municipality of Nova Iguaçu, State of Rio de Janeiro. Mem. Inst. Oswaldo Cruz 2002, 97, 799–800. [Google Scholar] [CrossRef]
- Peyrefitte, C.N.; Couissinier-Paris, P.; Mercier-Perennec, V.; Bessaud, M.; Martial, J.; Kenane, N.; Durand, J.P.A.; Tolou, H.J. Genetic Characterization of Newly Reintroduced Dengue Virus Type 3 in Martinique (French West Indies). J. Clin. Microbiol. 2003, 41, 5195–5198. [Google Scholar] [CrossRef]
- Ensafi, A.A.; Heydari-bafrooei, E.; Amini, M. DNA-functionalized biosensor for riboflavin based electrochemical interaction on pretreated pencil graphite electrode. Biosens. Bioelectron. 2012, 31, 376–381. [Google Scholar] [CrossRef]
- Hejazi, M.S.; Alipour, E.; Pournaghi-Azar, M.H. Immobilization and voltammetric detection of human interleukine-2 gene on the pencil graphite electrode. Talanta 2007, 71, 1734–1740. [Google Scholar] [CrossRef]
- Pournaghi-Azar, M.H.; Alipour, E.; Zununi, S. Direct and rapid electrochemical biosensing of the human interleukin-2 DNA in unpurified polymerase chain reaction (PCR)-amplified real samples. Biosens. Bioelectron. 2008, 24, 524–530. [Google Scholar] [CrossRef]
- Özcan, A.; Yücel, S. A novel approach for the determination of paracetamol based on the reduction of N-acetyl-p-benzoquinoneimine formed on the electrochemically treated pencil graphite electrode. Anal. Chim. Acta 2011, 685, 9–14. [Google Scholar] [CrossRef]
- Ermini, M.L.; Scarano, S.; Bini, R.; Banchelli, M.; Berti, D.; Mascini, M.; Minunni, M. A rational approach in probe design for nucleic acid-based biosensing. Biosens. Bioelectron. 2011, 26, 4785–4790. [Google Scholar] [CrossRef]
- Huang, J.; Yang, X.; He, X.; Wang, K.; Liu, J.; Shi, H.; Wang, Q.; Guo, Q.; He, D. Design and bioanalytical applications of DNA hairpin-based fluorescent probes. TrAC Trends Anal. Chem. 2014, 53, 11–20. [Google Scholar] [CrossRef]
- O’Brien, B.; Zeng, H.; Polyzos, A.A.; Lemke, K.H.; Weier, J.F.; Wang, M.; Zitzelsberger, H.F.; Weier, H.U.G. Bioinformatics tools allow targeted selection of chromosome enumeration probes and aneuploidy detection. J. Histochem. Cytochem. 2013, 61, 134–147. [Google Scholar] [CrossRef]
- Campos-Ferreira, D.S.; Souza, E.; Nascimento, G.; Zanforlin, D.; Arruda, M.; Beltrão, M.; Melo, A.; Bruneska, D.; Lima-Filho, J.L. Electrochemical DNA biosensor for the detection of human papillomavirus E6 gene inserted in recombinant plasmid. Arab. J. Chem. 2014. [Google Scholar] [CrossRef]
- Corrigan, D.K.; Schulze, H.; McDermott, R.A.; Schmüser, I.; Henihan, G.; Henry, J.B.; Bachmann, T.T.; Mount, A.R. Improving electrochemical biosensor performance by understanding the influence of target DNA length on assay sensitivity. J. Electroanal. Chem. 2014, 732, 25–29. [Google Scholar] [CrossRef]
- Soares, R.O.S.; Caliri, A. Stereochemical features of the envelope protein Domain III of dengue virus reveals putative antigenic site in the five-fold symmetry axis. Biochim. Biophys. Acta 2013, 1834, 221–230. [Google Scholar] [CrossRef]
- Weaver, S.C.; Brault, A.C.; Kang, W.; John, J. Genetic and Fitness Changes Accompanying Adaptation of an Arbovirus to Vertebrate and Invertebrate Cells Genetic and Fitness Changes Accompanying Adaptation of an Arbovirus to Vertebrate and Invertebrate Cells. J. Virol. 1999, 73, 4316–4326. [Google Scholar]
- Bennett, S.N.; Holmes, E.C.; Chirivella, M.; Rodriguez, D.M.; Beltran, M.; Vorndam, V.; Gubler, D.J.; McMillan, W.O. Molecular evolution of dengue 2 virus in Puerto Rico: Positive selection in the viral envelope accompanies clade reintroduction. J. Gen. Virol. 2006, 87, 885–893. [Google Scholar] [CrossRef]
- Weaver, S.C.; Vasilakis, N. Molecular evolution of dengue viruses: Contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol. 2009, 9, 523–540. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, Y.; Gao, F.; Jiang, S.; Zhang, B.; Ni, J.; Gao, F. A sensitive DNA biosensor based on a facile sulfamide coupling reaction for capture probe immobilization. Anal. Chim. Acta 2013, 788, 158–164. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Chen, M.; Luo, Y.; Deng, K.; Chen, D.; Fu, W. A new system for the amplification of biological signals: RecA and complimentary single strand DNA probes on a leaky surface acoustic wave biosensor. Biosens. Bioelectron. 2014, 60, 259–264. [Google Scholar] [CrossRef]
- Mohamadi, M.; Mostafavi, A.; Torkzadeh-Mahani, M. Electrochemical determination of biophenol oleuropein using a simple label-free DNA biosensor. Bioelectrochemistry 2015, 101C, 52–57. [Google Scholar] [CrossRef]
- Erdem, A.; Muti, M.; Karadeniz, H.; Congur, G.; Canavar, E. Colloids and Surfaces B: Biointerfaces Electrochemical monitoring of indicator-free DNA hybridization by carbon nanotubes—Chitosan modified disposable graphite sensors. Colloids Surf. B Biointerfaces 2012, 95, 222–228. [Google Scholar] [CrossRef]
- Paleček, E.; Fojta, M.; Tomschik, M.; Wang, J. Electrochemical biosensors for DNA hybridization and DNA damage. 1998; 13, 621–628. [Google Scholar]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–92. [Google Scholar] [CrossRef]
- Campos-Ferreira, D.S.; Nascimento, G.A.; Souza, E.V.M.; Souto-maior, M.A.; Arruda, M.S.; Zanforlin, D.M.L.; Ekert, M.H.F.; Bruneska, D.; Lima-filho, J.L. Electrochemical DNA biosensor for human papillomavirus 16 detection in real samples. Anal. Chim. Acta 2013, 804, 258–263. [Google Scholar] [CrossRef]
- Gao, Z.; Yang, W.; Wang, J.; Yan, H.; Yao, Y.; Ma, J.; Wang, B.; Zhang, M.; Liu, L. Electrochemical synthesis of layer-by-layer reduced graphene oxide sheets/polyaniline nanofibers composite and its electrochemical performance. Electrochim. Acta 2013, 91, 185–194. [Google Scholar] [CrossRef]
- Lucarelli, F.; Marrazza, G.; Palchetti, I.; Cesaretti, S.; Mascini, M. Coupling of an indicator-free electrochemical DNA biosensor with polymerase chain reaction for the detection of DNA sequences related to the apolipoprotein E. Anal. Chim. Acta 2002, 469, 93–99. [Google Scholar] [CrossRef]
- Teles, F.R.R.; Fonseca, L.P. Trends in DNA biosensors. Talanta 2008, 77, 606–623. [Google Scholar] [CrossRef]
- Tichoniuk, M.; Ligaj, M.; Filipiak, M. Application of DNA Hybridization Biosensor as a Screening Method for the Detection of Genetically Modified Food Components. Sensors 2008, 8, 2118–2135. [Google Scholar] [CrossRef]
- Liu, X.; Fan, Q.; Huang, W. DNA biosensors based on water-soluble conjugated polymers. Biosens. Bioelectron. 2011, 26, 2154–2164. [Google Scholar] [CrossRef]
- Gumustas, M.; Ozkan, S.A. The Role of and the Place of Method Validation in Drug Analysis Using Electroanalytical Techniques. Open Anal. Chem. J. 2011, 5, 1–21. [Google Scholar] [CrossRef]
- Nascimento, G.A.; Souza, E.V.M.; Campos-ferreira, D.S.; Arruda, M.S.; Castelletti, C.H.M.; Wanderley, M.S.O.; Ekert, M.H.F.; Bruneska, D. Electrochemical DNA biosensor for bovine papillomavirus detection using polymeric film on screen-printed electrode. Biosens. Bioelectron. 2012, 38, 61–66. [Google Scholar] [CrossRef]
- Aydoğdu, G.; Günendi, G.; Zeybek, D.K.; Zeybek, B.; Pekyardımcı, Ş. A novel electrochemical DNA biosensor based on poly-(5-amino-2-mercapto-1,3,4-thiadiazole) modified glassy carbon electrode for the determination of nitrofurantoin. Sens. Actuators B Chem. 2014, 197, 211–219. [Google Scholar]
- Wang, J.; Kawde, A. Pencil-based renewable biosensor for label-free electrochemical detection of DNA hybridization. Anal. Chim. Acta 2001, 431, 219–224. [Google Scholar] [CrossRef]
- Sehatnia, B.; Golabi, F.; Sabzi, R.E.; Hejazi, M.S. Modeling of DNA Hybridization Detection Using Methylene Blue as an Electroactive Label. J. Iran. Chem. Soc. 2011, 8, 115–122. [Google Scholar] [CrossRef]
- Souza, E.; Nascimento, G.; Santana, N.; Campos-ferreira, D.; Bibiano, J.; Arruda, M.S. Electrochemical DNA Biosensor for Sequences Related to the Human Papillomavirus Type 16 using Methylene Blue. Biosens. J. 2014, 3, 3–7. [Google Scholar]
- Ren, Y.; Deng, H.; Shen, W.; Gao, Z. A Highly Sensitive and Selective Electrochemical Biosensor for Direct Detection of MicroRNAs in Serum. Anal. Chem. 2013, 9, 4784–4789. [Google Scholar] [CrossRef]
- Auer, S.; Nirschl, M.; Schreiter, M.; Vikholm-Lundin, I. Detection of DNA hybridisation in a diluted serum matrix by surface plasmon resonance and film bulk acoustic resonators. Anal. Bioanal. Chem. 2011, 400, 1387–1396. [Google Scholar] [CrossRef]
- Gong, P.; Lee, C.; Gamble, L.J.; Castner, D.G.; Grainger, D.W.; Chem, L.J. A. Hybridization Behavior of Mixed DNA/Alkylthiol Monolayers on Gold: Characterization by Surface Plasmon Resonance and 32 P Radiometric Assay rescence intensity measurements reported in a related. 2006; 78, 3326–3334. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, N.; Souza, E.; Ferreira, D.; Zanforlin, D.; Bezerra, W.; Borba, M.A.; Arruda, M.; Lopes, K.; Nascimento, G.; Martins, D.; et al. A Sensitive and Selective Label-Free Electrochemical DNA Biosensor for the Detection of Specific Dengue Virus Serotype 3 Sequences. Sensors 2015, 15, 15562-15577. https://doi.org/10.3390/s150715562
Oliveira N, Souza E, Ferreira D, Zanforlin D, Bezerra W, Borba MA, Arruda M, Lopes K, Nascimento G, Martins D, et al. A Sensitive and Selective Label-Free Electrochemical DNA Biosensor for the Detection of Specific Dengue Virus Serotype 3 Sequences. Sensors. 2015; 15(7):15562-15577. https://doi.org/10.3390/s150715562
Chicago/Turabian StyleOliveira, Natália, Elaine Souza, Danielly Ferreira, Deborah Zanforlin, Wessulla Bezerra, Maria Amélia Borba, Mariana Arruda, Kennya Lopes, Gustavo Nascimento, Danyelly Martins, and et al. 2015. "A Sensitive and Selective Label-Free Electrochemical DNA Biosensor for the Detection of Specific Dengue Virus Serotype 3 Sequences" Sensors 15, no. 7: 15562-15577. https://doi.org/10.3390/s150715562
APA StyleOliveira, N., Souza, E., Ferreira, D., Zanforlin, D., Bezerra, W., Borba, M. A., Arruda, M., Lopes, K., Nascimento, G., Martins, D., Cordeiro, M., & Lima-Filho, J. (2015). A Sensitive and Selective Label-Free Electrochemical DNA Biosensor for the Detection of Specific Dengue Virus Serotype 3 Sequences. Sensors, 15(7), 15562-15577. https://doi.org/10.3390/s150715562





