Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological and Chemical Materials
2.2. Bacterial Strains
2.3. Preparation of DNA Library
2.4. PCR Optimization
2.5. SELEX Procedures
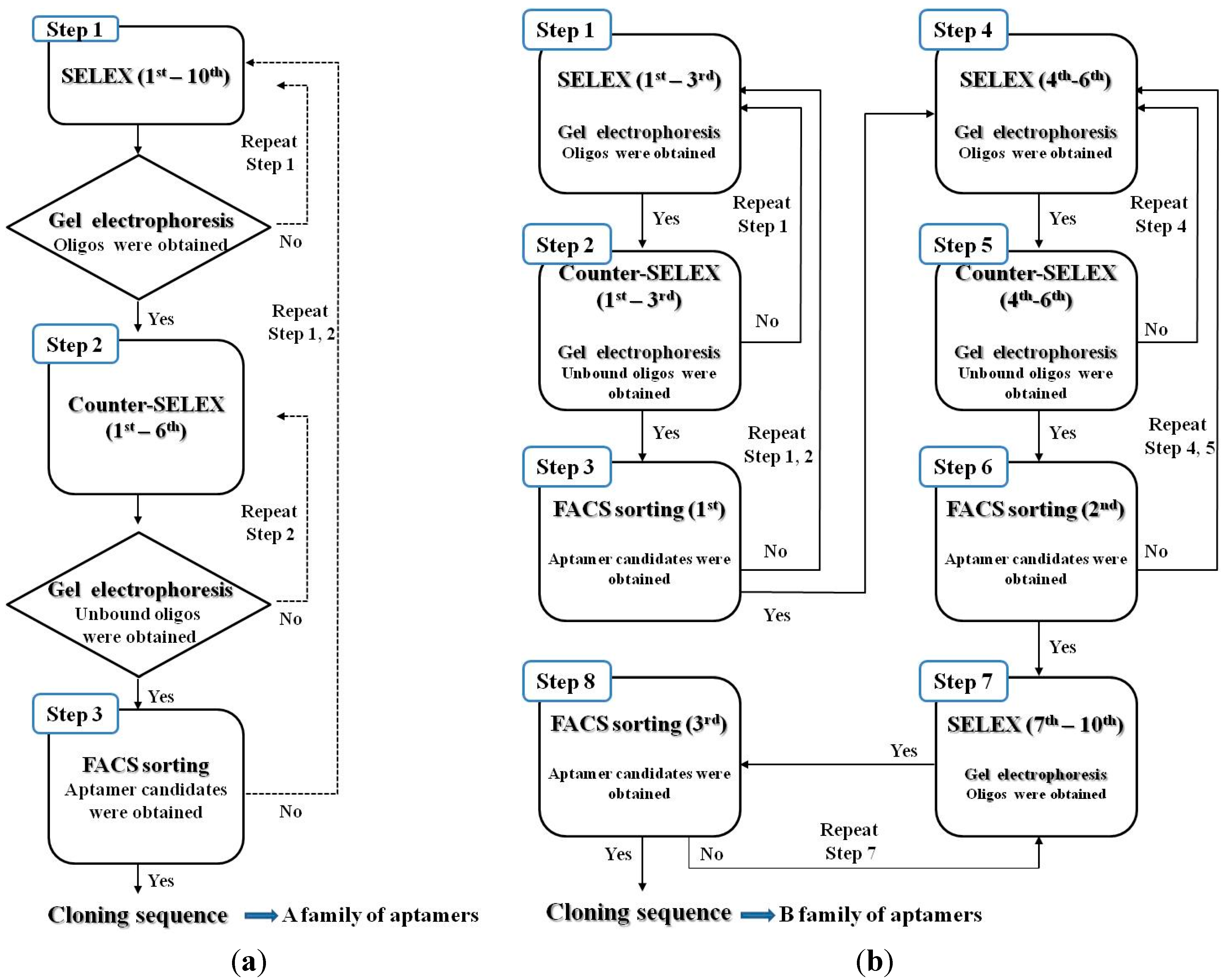
2.6. Fluorescence-Activated Cell Sorting (FACS)
2.7. Characterization of Binding Parameters of Aptamers to Bacteria
2.8. Aptamer-Binding Assays and Prediction of Candidate Aptamer Sequences
3. Results and Discussion
3.1. Screening of Aptamer Candidates in the Basic Whole-Cell SELEX
| Aptamers | Sequences |
|---|---|
| A2 | ACGGGCGTGGGAGGCAATGCCTTGCTTGTAGGCTTCCCCTGTGCGCG |
| A14 | CACACCGCAGCAGTGGGAACGTTTCAGCCATGCAAGCATCACGCCCGT |
| A15 | CACGCGCAAACAGATTAACACTCCGCCTAAGTCTGCCGCACGC |
| A20 | GCGTGCAGCGGGGGCTGCGCGGTGGAGTGCTGTGGGCG |
| B3 | GCGTGCGGAGCCAGGATGGGAGGTCTGTAGGTCTGCGGGGCGTG |
| B6 | GCGTGTCGGTGTCTGCCGGGGGATGTGGAGGCTGGGTGTTGCGCG |
| B7 | GCGTGGGCGGGCTACCTGGCTAGTACGCCATGATGCCTGCACGCG |
| B15 | CACGCGCAAACAGATTAACACTCCGCCTAAGTCTGCCGCACGC |
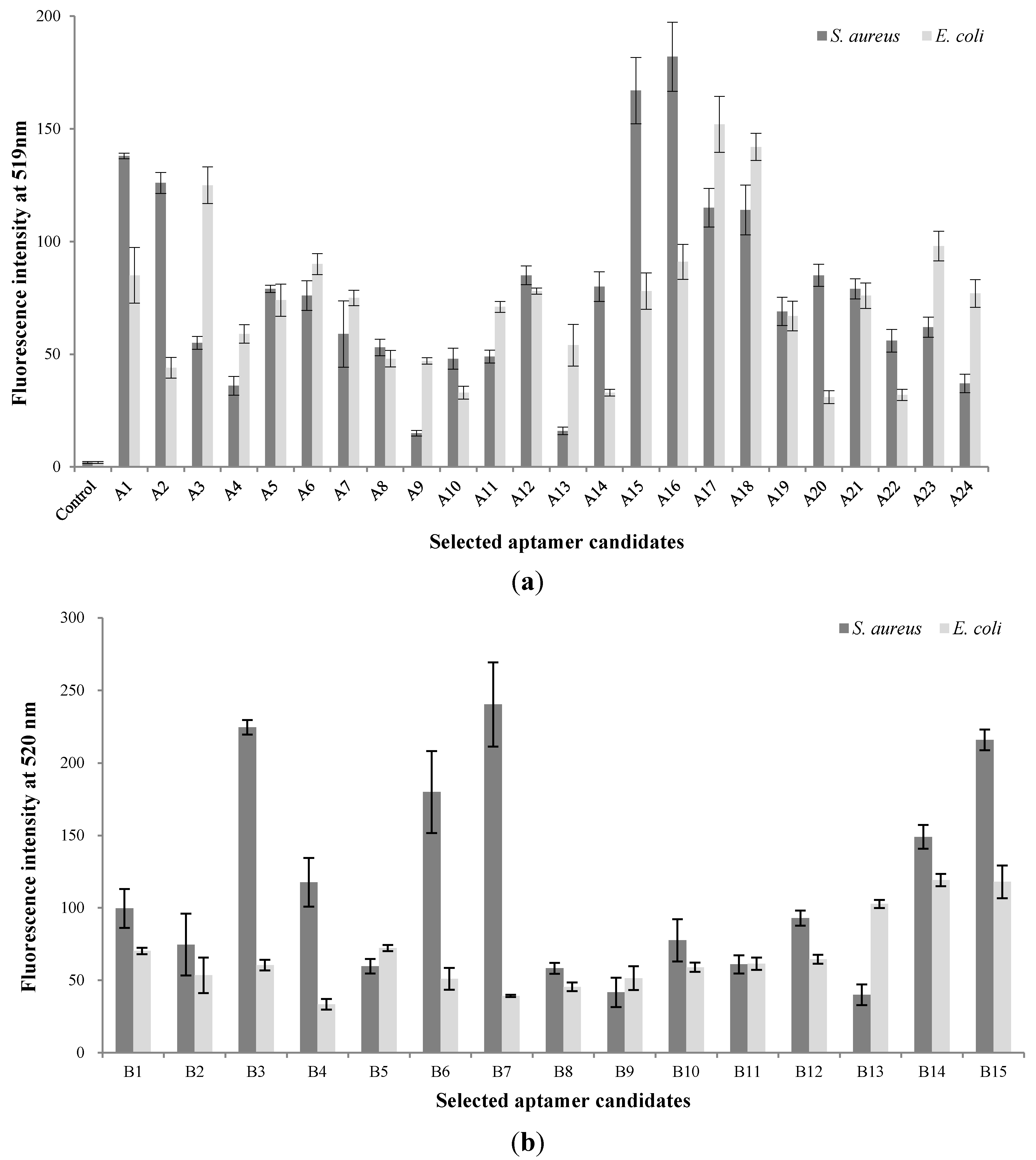
3.2. Screening of Aptamer Candidates in the Modified Whole-Cell SELEX
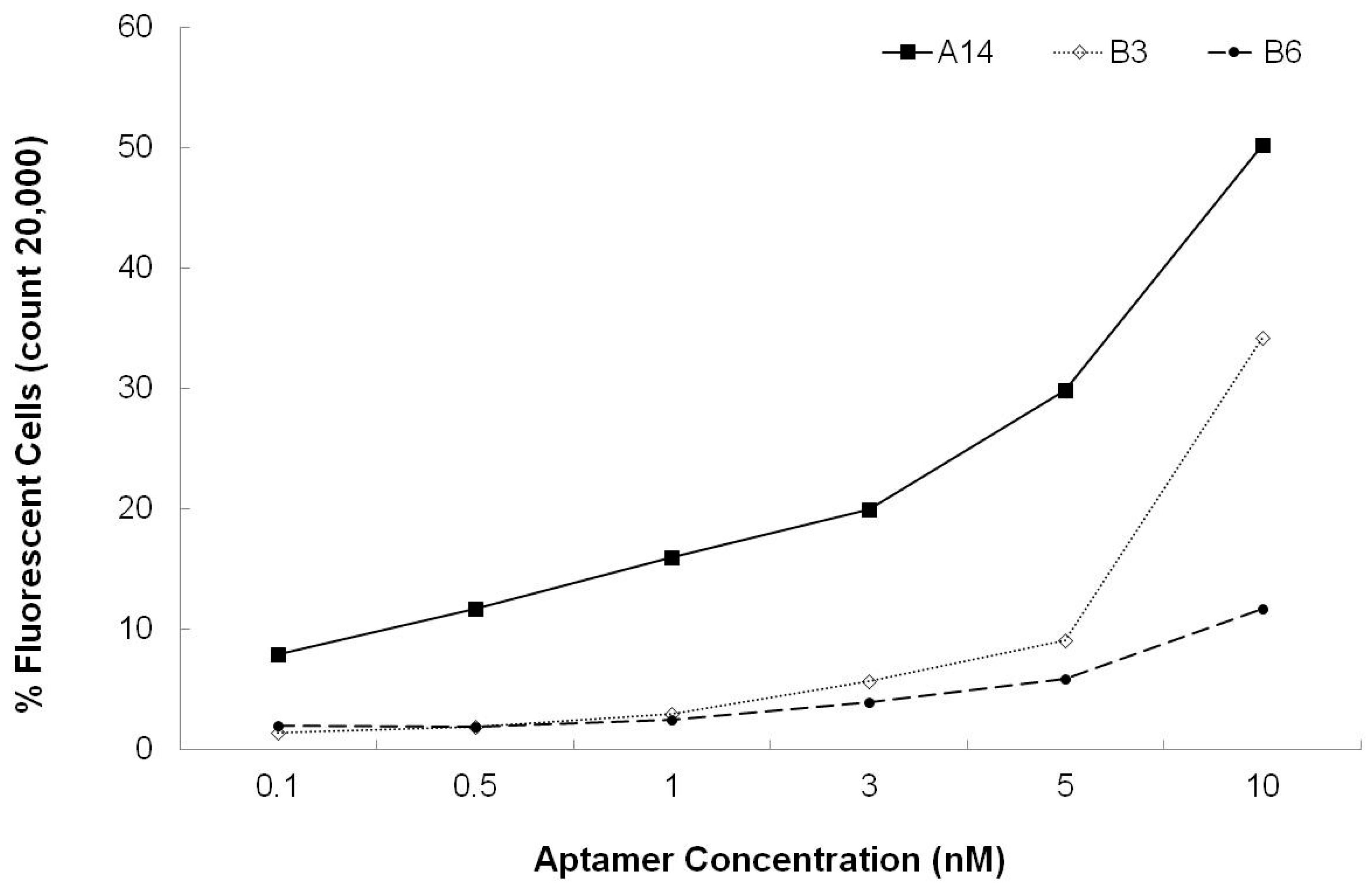
3.3. Dissociation Constants for Aptamers

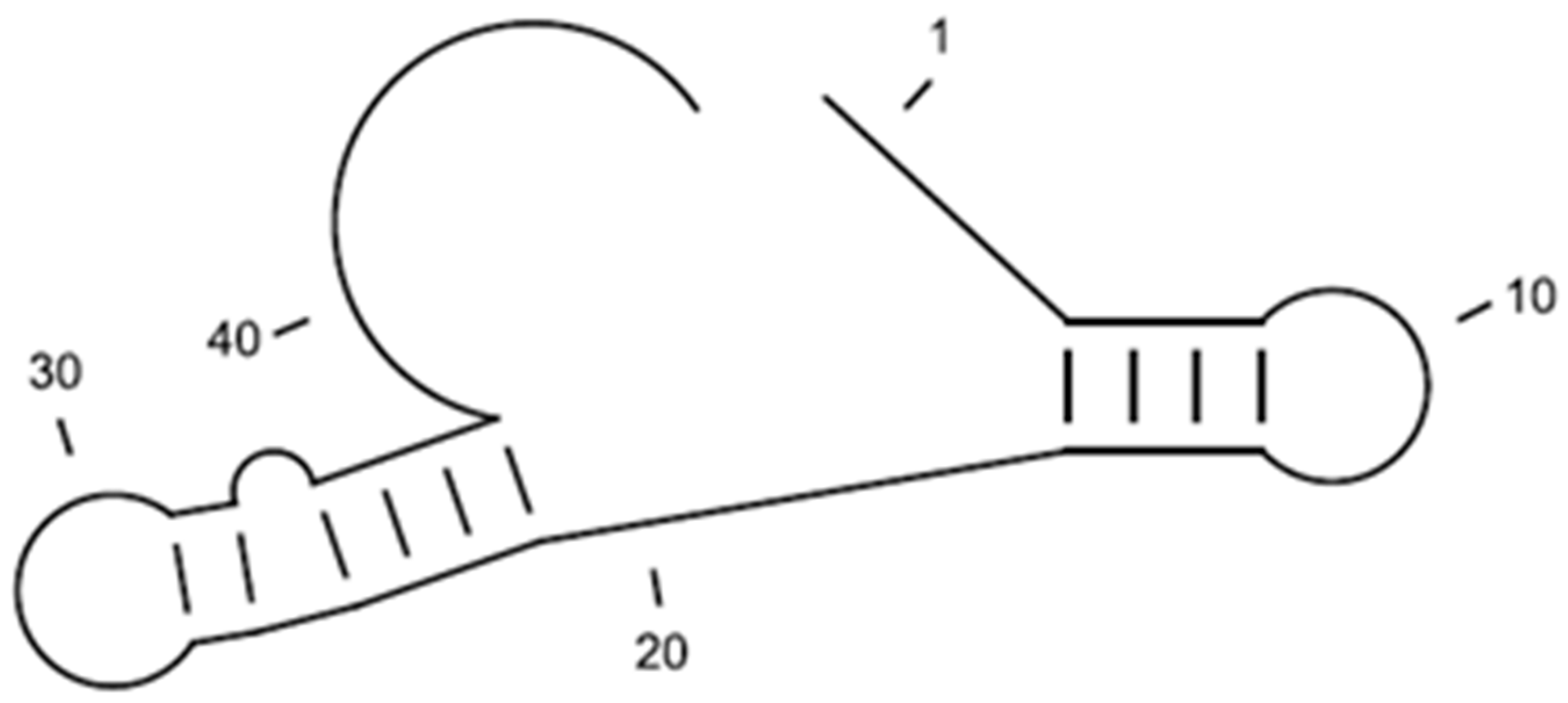
4. Conclusions/Outlook
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Loir, Y.L.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Gen. Mol. Res. 2003, 2, 63–76. [Google Scholar]
- Lowy, F.D. Staphylococcus aureus infections. New Engl. J. Med. 1998, 8, 520–532. [Google Scholar] [CrossRef]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Han, S.R.; Lee, S.W. In vitro selection of RNA aptamer specific to Staphylococcus aureus. Ann. Microbiol. 2014, 64, 883–885. [Google Scholar] [CrossRef]
- Food Poisoning Statistics in Korea; Ministry of Food and Drug Safety in Korea: Chungcheongbuk-do, Korea. Available online: http://www.mfds.go.kr (accessed on 1 July 2014).
- Foodborne Pathogenic Microorganisms and Natural Toxins: Bad Bug Book; US Food and Drug Administration: Silver Spring, MD, USA. Available online: http://www.fda.gov (accessed on 1 July 2014).
- Duan, N.; Wu, S.; Zhu, C.; Ma, X.; Wang, Z.; Yu, Y.; Jiang, Y. Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Anal. Chim. Acta 2012, 723, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Moon, J.H.; Hahm, B.K.; Morgan, M.; Bhunia, A.; Om, A.S. Rapid detection of Salmonella enteritidis in pork samples with impedimetric biosensor: Effect of electrode spacing on sensitivity. Food Sci. Biotechnol. 2009, 18, 89–94. [Google Scholar]
- Kim, G.; Moon, J.H.; Morgan, M. Multivariate data analysis of impedimetric biosensor responses from Salmonella typhimurium. Anal. Methods 2013, 5, 4074–4080. [Google Scholar] [CrossRef]
- Arora, P; Sindhu, A.; Kaur, H.; Dilbaghi, N.; Chauhury, A. An overview of transducers as platform for the rapid detection of foodborne pathogens. Appl. Microbiol. Biotechnol. 2013, 97, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Cullum, B. Biosensors and biochips: Advances in biological and medical diagnostics. Fresenius J. Anal. Chem. 2000, 366, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S. Cell-SELEX technology. Biores. Open Access 2012, 1, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Medley, C.D.; Bamrungsap, S.; Tan, W.; Smith, J.E. Aptamer-conjugated nanoparticles for cancer cell detection. Anal. Chem. 2011, 83, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Torres-Chavolla, E.E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectronics 2009, 24, 3175–3182. [Google Scholar] [CrossRef]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.A. Selection of DNA aptamers for capture and detection of Salmonella typhimurium using a whole-cell SELEX approach in conjunction with cell sorting. Appl. Microbiol. Biotechnol. 2013, 97, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, S.; Chen, L.; Ding, H.; Xu, H.; Huang, Y.; Li, J.; Liu, N.; Cao, W.; Zhu, Y.; et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Ahmed, M.S.L.; Dolf, A.; Endl, E.; Knolle, P.A.; Fumulok, M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010, 5, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Cibiel, A.; Dupont, D.M.; Ducongé, F. Methods to identify aptamers against cell surface biomarkers. Pharmaceuticals 2011, 4, 1216–1235. [Google Scholar] [CrossRef]
- Moon, J.; Kim, G.; Lee, S.; Park, S. Identification of Salmonella typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J. Microbiol. Methods 2013, 95, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.A.; Zhang, H.; Guan, L.L.; Li, X.F.; Le, X.C. Selection of aptamers against live bacterial cells. Anal. Chem. 2008, 80, 7812–7819. [Google Scholar] [CrossRef] [PubMed]
- Houston, P.; Kodadek, T. Spectrophotometric assay for enzyme-mediated unwinding of double-stranded DNA. Proc. Natl. Acad. Sci. USA 1994, 91, 5471–5474. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Yang, C.Y.; Sun, R.L.; Cheng, Y.F.; Kao, W.C.; Yang, P.C. Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles. Sci. Rep. 2013. [Google Scholar] [CrossRef]
- Suh, S.H.; Dwivedi, H.P.; Choi, S.J.; Jaykus, L.A. Selection and characterization of DNA aptamers specific for Listeria species. Anal. Biochem. 2014, 459, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 2010, 87, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.Y.; Chon, J.W.; Choi, I.S.; Park, C.; Kim, D.E.; Seo, K.H. Development of RNA aptamers for detection of Salmonella Enteritidis. J. Microbiol. Methods 2012, 87, 79–82. [Google Scholar] [CrossRef]
- Joshi, R.; Janagama, H.; Dwivedi, H.P.; Kumar, T.M.A.S.; Jaykus, L.A. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes 2009, 23, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Han, S.Y.; Maeng, J.S.; Cho, Y.J.; Lee, S.W. In vitro selection of Escherichia coli O157:H7-specific RNA aptamer. Biochem. Bioph. Res. Commun. 2011, 417, 414–420. [Google Scholar] [CrossRef]
- Meyer, M.; Scheper, T.; Walter, J.G. Aptamers: Versatile probes for flow cytometry. Appl. Microbiol. Biotechnol. 2013, 97, 7097–7109. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Maufort, J.P.; Nie, J.; Stewart, R.; Mclntosh, B.E.; Conti, L.R.; Ahmad, K.M.; Soh, H.T.; Thomson, J.A. Development of an efficient targeted cell-SELEX procedure for DNA aptamer reagents. PLoS ONE 2013, 8, e71798. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, J.; Kim, G.; Park, S.B.; Lim, J.; Mo, C. Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers. Sensors 2015, 15, 8884-8897. https://doi.org/10.3390/s150408884
Moon J, Kim G, Park SB, Lim J, Mo C. Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers. Sensors. 2015; 15(4):8884-8897. https://doi.org/10.3390/s150408884
Chicago/Turabian StyleMoon, Jihea, Giyoung Kim, Saet Byeol Park, Jongguk Lim, and Changyeun Mo. 2015. "Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers" Sensors 15, no. 4: 8884-8897. https://doi.org/10.3390/s150408884
APA StyleMoon, J., Kim, G., Park, S. B., Lim, J., & Mo, C. (2015). Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers. Sensors, 15(4), 8884-8897. https://doi.org/10.3390/s150408884






