Microfluidic Organ/Body-on-a-Chip Devices at the Convergence of Biology and Microengineering
Abstract
:1. Introduction
2. Microfluidics—from Small Benchtop Biosensors to High-Throughput Systems
3. Convergence between Microfluidics and Tissue Engineering: Bio-MEMS and Organ-on-a-Chip
| Application | Platform | References |
|---|---|---|
| Cardiovascular System | ||
| Angiogenesis studies | Dual channel chip/angiogenesis model, microfluidic tri-culture platform | [58,59] |
| Biophysical studies | Pressure attenuator + Funnel chain/cell deformability microfluidic device | [60] |
| Muscular thin films | [61] | |
| Microfluidics + optical microscopy | [62] | |
| Microfluidics + ultrasound imaging system | [63] | |
| High-speed video microscopy in microcapillaries | [64] | |
| Drug screening/development | Microchannel microfluidic chip | [65] |
| Laminar ventricular muscle-on-a-chip | [66] | |
| Organ/tissue structure/activity | Microfluidic cardiac cell culture model, heart-on-a-chip, artery-on-a-chip, microscale blood vessel module (µBVM) in a single microchannel device, microfluidic perfusion cell culture chip, microfluidic delivery system, microchannel biochips as vaso-occlusive processes model, perfusion microfluidic device, branched microfluidic channels | [61,67,68,69,70,71,72,73,74] |
| Respiratory system | ||
| Biological barriers | Flow stretch chip | [75] |
| Compartmentalized microwells in a microfluidic device | [76] | |
| Cancer mechanisms | Microfluidics + electric fields | [77] |
| Cell culture | Biomimetic microfluidic airway model | [78] |
| Cell differentiation | 3D gelatin-microbbuble scaffold produced by microfluidic device | [79] |
| Cell migration | Dynamic transwell microfluidic system + perfusion culture, microfluidic gradient generator | [80,81] |
| Drug delivery | Microfludics + surface acoustic wave (SAW) nebulizer | [82] |
| In vivo organ studies | Microfludics + single oxygenator units | [83] |
| Molecular mechanisms | Microfluidics + concentration gradient generator | [84] |
| Wound healing | Microfluidic system of converging multichannels + hydrodynamic flow focusing | [85] |
| Nervous System | ||
| Axonal transport | Microchannels/microgrooves + compartmented microfluidic culture | [86,87,88,89] |
| Cell culture | Microchannels/microgrooves + compartmented microfluidic co-cultures, shear-free microfluidic gradient generator | [90,91] |
| Cell line characterization | Microfluidics + electrophoresis | [92] |
| Microfluidics + quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) | [93] | |
| Cell differentiation | Microgrooves + neuronal compartment + myelination compartment microfluidic co-cultures | [94] |
| Cell migration | Microfluidic microgrooves + compartment to culture explants + compartment with Matrigel® to receive migrating neurons | [95] |
| Cellular/Molecular mechanisms | Two-compartment microfluidic culture system (neuronal compartment + myelination compartment) microfluidic co-cultures, microfluidic axon-microglia platform, axon injury micro-compression platform | [94,96,97,98] |
| Microfluidic devices or bioreactors + ultra-performance liquid chromatography-ion mobility-mass (UPLC-IM-MS) | [99] | |
| Drug delivery | Microfluidic + perfusion device | [100] |
| Drug screening/development | Microfluidic “Fish-Trap” array, gravity-induced flow + microfluidic chip | [101,102] |
| Microfluidics + trans-endothelial electrical resistance (TEER) | [103] | |
| Organ/tissue structure/activity | Microfluidic “Fish-Trap” array, two-compartment + microchannels microfluidic culture system | [90,101] |
| Screening / Diagnostic | Microfluidic cell sorter | [104] |
| Synaptic studies | Three compartment microfluidic device competition experiment, two cell culture chambers + funnel-shaped micro-channels microfluidic device | [105,106] |
| Toxicity studies | Axonal microfluidic chambers | [107] |
| Microfluidics + 96-well plate | [108] | |
| Digestive + Excretory System | ||
| Cell culture | Biomimic hydrogel nephron | [109] |
| Integrated Dynamic Cell Culture Microchip (IDCCM), Microfluidic endothelial-like barrier, dam-wall and nozzle microfluidic device, hemi-coaxial-flow channel microfluidic, dual perifusion platform | [110,111,112,113,114] | |
| Microfluidic bioreactor | [115,116,117] | |
| Microfluidic droplet-based cell encapsulation | [118] | |
| Multiwell culture system | [119,120] | |
| Microfluidic-multilayer device (MMD) | [121] | |
| Cell differentiation | Microfluidic cell culture chamber/channels | [122,123] |
| Microfluidics + qRT-PCR | [124] | |
| Circulating tumor cells studies | Microfluidic geometrically enhanced mixing chip, Geometrically Enhanced Differential Immunocapture (GEDI) device | [125,126,127] |
| Drug screening/development | Gut-on-a-chip, 3D villi scaffold + microfluidic device, IDCCM | [128,129,130,131] |
| Microfluidics + optical fiber | [132] | |
| Microfluidic cell culture array | [133] | |
| Microfluidic droplet-based cell encapsulation | [118] | |
| Three-dimensional microfluidic microanalytical micro-organ device (3MD) | [134,135] | |
| Food analysis | Microfluidics + Fluorescence imaging | [136] |
| Metabolism studies | IDCCM, two-plate bioreactor, metabolomics-on-a-chip, microfluidic delivery device, two-color detection microfluidic system, multimodal islet hypoxia device | [110,117,131,137,138,139,140] |
| Microfluidic bioreactor | [141] | |
| Microscale cell culture analogue (μCCA) | [142] | |
| Microfluidics-optical sensor | [143] | |
| Multiwell culture system | [119] | |
| Organ-organ interaction | Integrated Insert in a Dynamic Microfluidic Platform (IIDMP), on-chip small intestine-liver coupled microfluidic network | [144,145] |
| Screening/Diagnostic | Microfluidics + surface plasmon resonance | [146] |
| Microfluidics + optoelectronic sensor | [147] | |
| Microfluidics + optomechanical metric | [148] | |
| Therapeutic systems | Wearable ultrafiltration units for dialysis | [149,150] |
| Toxicity studies | Metabolomics-on-a-chip, Gut-on-a-chip, IDDCM bioreactor, pharmacokinetic microfluidic perfusion system | [137,151,152,153,154] |
| Kidney and kidney/liver microfluidic biochips | [155,156,157] | |
| Microfluidics + optical fiber | [132] | |
| μCCA | [142,158] | |
| Microfluidic bioreactor | [159] | |
| Microfluidic human kidney proximal tubule-on-a-chip device | [160] | |
| MMD | [121] | |
| Multiwell culture system | [119] | |
| Endocrine System | ||
| Cancer mechanisms | Microfluidic co-culture model, chemokine gradient + 3D culture device | [161,162] |
| Fertilization | Motile spermatozoa sorter + microfluidic chip, microfluidic device mimicking female reproductive tract | [163,164] |
| Metabolism studies | Microfluidics + resonant waveguide grating (RWG) sensor | [165] |
| Monitoring | Microfluidics + electrochemical sensor | [166] |
| Screening and diagnostic | Blood plasma separation microfluidic chip | [18] |
| Microfluidics + optical sensor | [167] | |
| Microfluidics + liquid chromatography-mass spectrometry | [168,169] | |
| Microfluidics + potentiostat | [170] | |
| Microfluidics + electrochemical sensor | [171] | |
| Digital microfluidics | [172] | |
| Integumentary System | ||
| Biological barriers | Stable gel/liquid interface microfluidic chip | [173] |
| Cell differentiation | Pillar array microfluidic device based on cell surface markers | [174] |
| Cell migration | 3D matrices microfluidic device | [175] |
| Screening and diagnostic | Microfluidics + conductometric sensor | [176] |
| Microfluidics + potentiometric sensor | [177] | |
| Skin repair | Microfluidic wound-healing model + wound dressing screening | [178,179] |
3.1. Cardiovascular System
3.2. Respiratory System
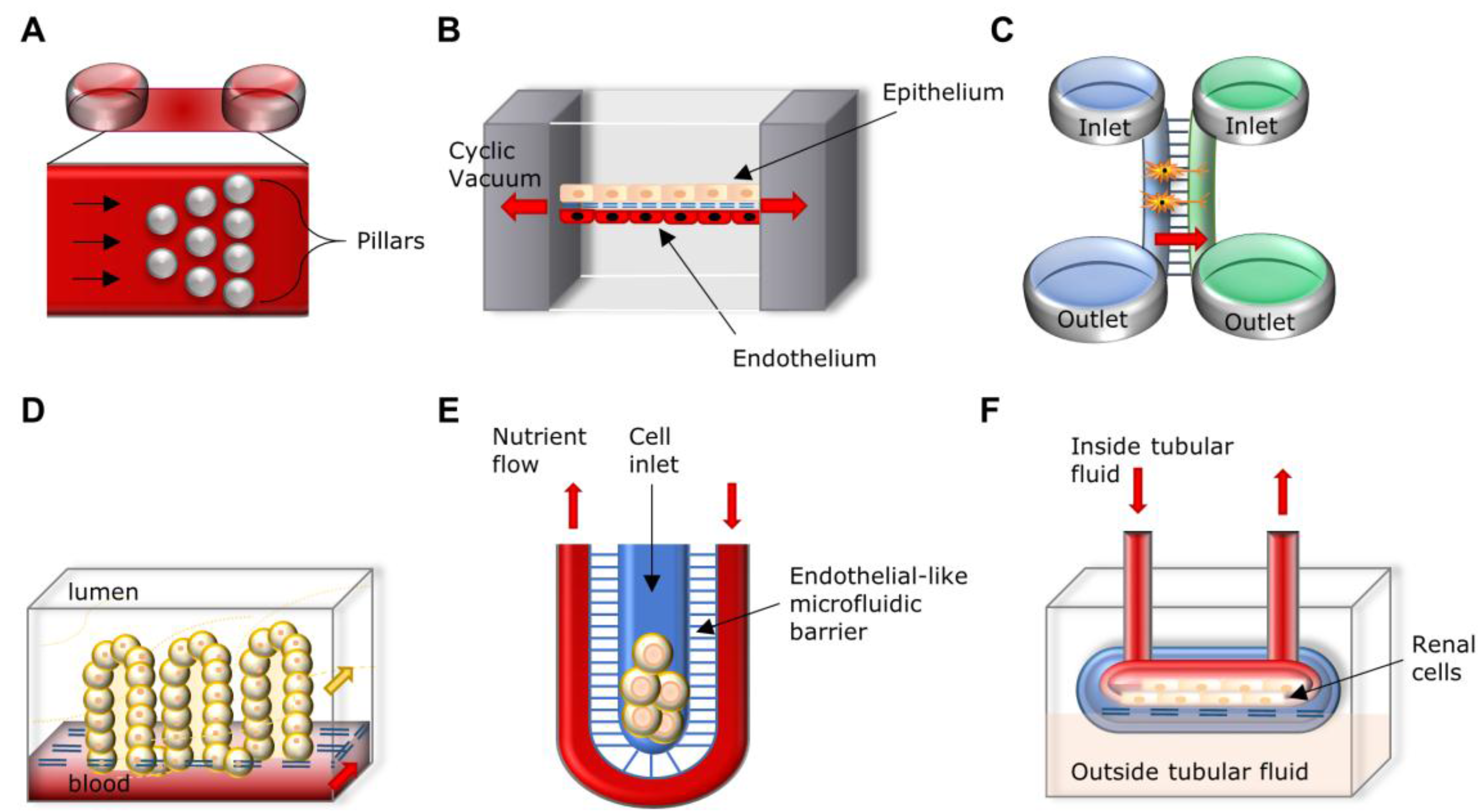
3.3. Nervous System
3.4. Digestive and Excretory Systems
3.5. Other Promising Applications for Microfluidics Technology
4. Body-on-a-Chip: A Future Perspective
- high accuracy prediction and comprehensive analyses of novel therapeutic candidates during preclinical stages, by a closer estimation of efficacy and dose response;
- reduction and likely replacement of animals in preclinical drug development, thereby reducing costs and time to market;
- creation of a drug development tool that helps modern medicine rapidly respond to fast-moving pandemics or chemical warfare/bioterrorism attacks;
- study cell signaling by monitoring the metabolites that are consumed, produced, and exchanged between all tissues at physiologically relevant concentrations in real time;
- study embryology and its signaling pathways by following intercellular signals and/or bioelectrical messages;
- conduct experiments that cannot be performed in cell culture, e.g., study of tissue-tissue interactions that occur as a result of metabolite travelling from one tissue to other distant tissue, and through dynamic forces that resemble blood circulation;
- efficient and reliable cell–cell and cell-drug/biomaterial interaction studies, narrowing the gap between in vivo and in vitro conditions.
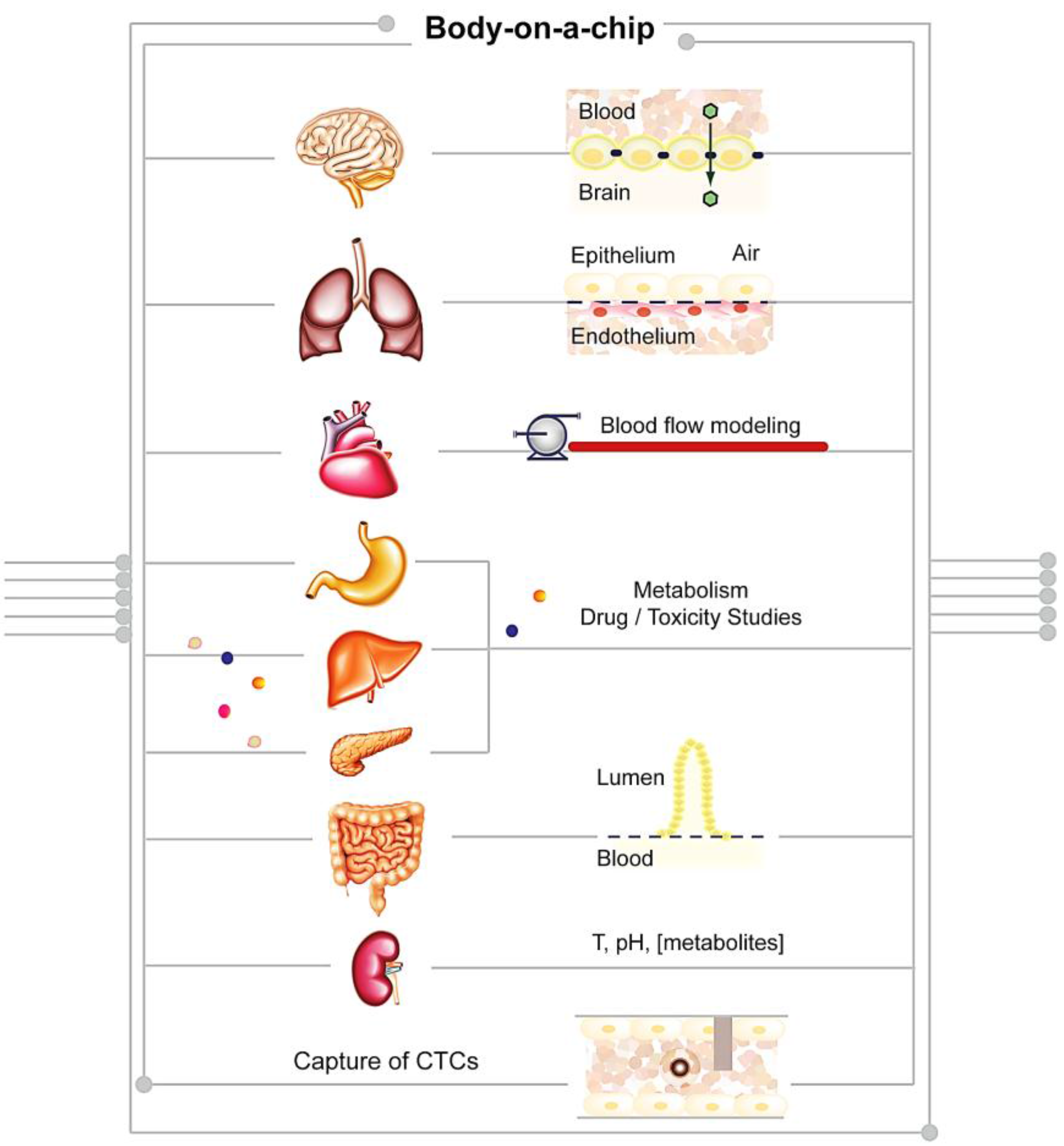
4.1. Proposed Applications of BoCs
| Organs / Interactions | Device / Platform Name | Application | References |
|---|---|---|---|
| Brain, Heart, Lung, Skin, Adipose, Muscle, Liver, Bone Marrow, Kidney | Physiologically-based pharmacokinetic (PBPK) model | ADME profiling and quantification of the amount of drugs in different parts of the body | [212,213] |
| Gastrointestinal Tract and Liver | µCCA | Evaluating nanoparticle toxicity and interactions with tissues | [200] |
| Gut—parallel tube model | Investigate paracetamol intestinal and liver first pass metabolism | [206] | |
| Heart, Liver, Vascular System | HeLiVa | Drug testing in human health and disease | [205] |
| Intestine, Liver, Skin and Kidney | Four-Organ-Chip | ADME profiling and toxicity testing | [207] |
| Liver, Colorectal Tissues | 96-well format-based microfluidic platform | Testing drug effects at different concentrations in several tissues | [209,210,211] |
| Liver, Heart, Lung and Kidney | ATHENA (“Homo Minutus”) | Screening new drugs for potency and potential side-effects | [214] |
| Liver, Tumor and Marrow | Pharmacokinetic-pharmacodynamic (PK-PD) model combined with a µCCA | Testing drug toxicity and improve insights into the drug’s mechanism of action | [212] |
| Lung, Gut | PDMS-based organs-on-chip | Prediction of clinical responses in humans | [152,215] |
4.2. BoCs and Cancer
4.3. Limitations of BoCs
5. Concluding Remarks
Acknowledgments
Conflicts of Interest
List of Abbreviations
References
- Langer, R.; Vacanti, J.P. Tissue Engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic organ-on-a-chip technology for advancement of drug development and toxicology. Adv. Healthc. Mater. 2015, 4, 1426–1450. [Google Scholar] [CrossRef] [PubMed]
- Esch, E.W.; Bahinski, A.; Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015, 14, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Skommer, J.; Wlodkowic, D. Successes and future outlook for microfluidics-based cardiovascular drug discovery. Exp. Opin. Drug Discov. 2015, 10, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Esteves-Villanueva, J.O.; Trzeciakiewicz, H.; Martic, S. A protein-based electrochemical biosensor for detection of tau protein, a neurodegenerative disease biomarker. Analyst 2014, 139, 2823–2831. [Google Scholar] [CrossRef] [PubMed]
- Vistas, C.R.; Soares, S.S.; Rodrigues, R.M.M.; Chu, V.; Conde, J.P.; Ferreira, G.N.M. An amorphous silicon photodiode microfluidic chip to detect nanomolar quantities of HIV-1 virion infectivity factor. Analyst 2014, 139, 3709–3713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yin, B.-C.; Wang, X.-F.; Ye, B.-C. Interaction of peptides with graphene oxide and its application for real-time monitoring of protease activity. Chem. Commun. 2011, 47, 2399–2401. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Chirumamilla, M.; Toma, A.; Gopalakrishnan, A.; Zaccaria, R.P.; Alabastri, A.; Leoncini, M.; Di Fabrizio, E. Plasmon based biosensor for distinguishing different peptides mutation states. Sci. Rep. 2013, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.J.; Chen, H.Y. Amperometric glucose sensor based on coimmobilization of glucose oxidase and poly(p-phenylenediamine) at a platinum microdisk electrode. Anal. Biochem. 2000, 280, 221–226. [Google Scholar]
- Mishra, G.K.; Sharma, A.; Deshpande, K.; Bhand, S. Flow injection analysis biosensor for urea analysis in urine using enzyme thermistor. Appl. Biochem. Biotechnol. 2014, 174, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Javanmard, M.; Davis, R.W. A microfluidic platform for electrical detection of DNA hybridization. Sens. Actuators B Chem. 2011, 154, 22–27. [Google Scholar] [CrossRef] [PubMed]
- De-Carvalho, J.; Rodrigues, R.M.M.; Tomé, B.; Henriques, S.F.; Mira, N.P.; Sá-Correia, I.; Ferreira, G.N.M. Conformational and mechanical changes of DNA upon transcription factor binding detected by a QCM and transmission line model. Analyst 2014, 139, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Yin, T.-I.; Reyes, D.; Urban, G.A. A microfluidic chip with integrated electrical cell-impedance sensing for monitoring single cancer cell migration in three-dimensional matrixes. Anal. Chem. 2013, 85, 11068–11076. [Google Scholar] [CrossRef] [PubMed]
- Weltin, A.; Slotwinski, K.; Kieninger, J.; Moser, I.; Jobst, G.; Wego, M.; Ehret, R.; Urban, G.A. Cell culture monitoring for drug screening and cancer research: A transparent, microfluidic, multi-sensor microsystem. Lab Chip 2014, 14, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Liu, J.; Dvir, T.; Jin, L.; Tsui, J.H.; Qing, Q.; Suo, Z.; Langer, R.; Kohane, D.S.; Lieber, C.M. Macroporous nanowire nanoelectronic scaffolds for synthetic tissues. Nat. Mater. 2012, 11, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Manz, A.; Harrison, D.J.; Verpoorte, E.M.J.; Fettinger, J.C.; Paulus, A.; Lüdi, H.; Widmer, H.M. Planar chips technology for miniaturization and integration of separation techniques into monitoring systems—Capillary electrophoresis on a chip. J. Chromatogr. A 1992, 593, 253–258. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Madadi, H.; Casals-Terré, J.; Mohammadi, M. Self-driven filter-based blood plasma separator microfluidic chip for point-of-care testing. Biofabrication 2015, 7, 025007:1–025007:11. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.M.; Liu, A.P. The application of micropipette aspiration in molecular mechanics of single cells. J. Nanotechnol. Eng. Med. 2014, 5, 040801:1–040801:6. [Google Scholar] [CrossRef] [PubMed]
- Giobbe, G.G.; Michielin, F.; Luni, C.; Giulitti, S.; Martewicz, S.; Dupont, S.; Floreani, A.; Elvassore, N. Functional differentiation of human pluripotent stem cells on a chip. Nat. Methods 2015, 12, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Vasiliauskas, R.; Liu, D.; Cito, S.; Zhang, H.; Shahbazi, M.-A.; Sikanen, T.; Mazutis, L.; Santos, H.A. Simple microfluidic approach to fabricate monodisperse hollow microparticles for multidrug delivery. ACS Appl. Mater. Interfaces 2015, 7, 14822–14832. [Google Scholar] [CrossRef] [PubMed]
- Pullagurla, S.R.; Witek, M.A.; Jackson, J.M.; Lindell, M.A.M.; Hupert, M.L.; Nesterova, I.V.; Baird, A.E.; Soper, S.A. Parallel affinity-based isolation of leukocyte subsets using microfluidics: Application for stroke diagnosis. Anal. Chem. 2014, 86, 4058–4065. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.S.Y.; Chua, C.; Gole, L.; Biswas, A.; Koay, E.; Choolani, M. Same-day prenatal diagnosis of common chromosomal aneuploidies using microfluidics-fluorescence in situ hybridization. Prenat. Diagn. 2012, 32, 321–328. [Google Scholar] [CrossRef] [PubMed]
- De la Rica, R.; Stevens, M.M. Plasmonic ELISA for the detection of analytes at ultralow concentrations with the naked eye. Nat. Protoc. 2013, 8, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Stern, E.; Vacic, A.; Rajan, N.K.; Criscione, J.M.; Park, J.; Ilic, B.R.; Mooney, D.J.; Reed, M.A.; Fahmy, T.M. Label-free biomarker detection from whole blood. Nat. Nanotechnol. 2010, 5, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Andreou, C.; Hoonejani, M.R.; Barmi, M.R.; Moskovits, M.; Meinhart, C.D. Rapid detection of drugs of abuse in saliva using surface enhanced raman spectroscopy and microfluidics. ACS Nano 2013, 7, 7157–7164. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Leung, K.W.; Ting, A.K.L.; Wong, Z.C.F.; Ng, W.Y.Y.; Choi, R.C.Y.; Dong, T.T.X.; Wang, T.; Lau, D.T.W.; Tsim, K.W.K. Microfluidic chip based nano liquid chromatography coupled to tandem mass spectrometry for the determination of abused drugs and metabolites in human hair. Anal. Bioanal. Chem. 2012, 402, 2805–2815. [Google Scholar] [CrossRef] [PubMed]
- Swensen, J.S.; Xiao, Y.; Ferguson, B.S.; Lubin, A.A.; Lai, R.Y.; Heeger, A.J.; Plaxco, K.W.; Soh, H.T. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J. Am. Chem. Soc. 2009, 131, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.C.; Hanes, R.D. A microfluidic device for presumptive testing of controlled substances. J. Forensic Sci. 2007, 52, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, J.; Song, Y.; Xu, Y.; Pan, X.; Sun, Y.; Li, D. A label-free microfluidic biosensor for activity detection of single microalgae cells based on chlorophyll fluorescence. Sensors 2013, 13, 16075–16089. [Google Scholar] [CrossRef] [PubMed]
- Buffi, N.; Merulla, D.; Beutier, J.; Barbaud, F.; Beggah, S.; van Linte, H.; Renaud, P.; van der Meer, J.R. Miniaturized bacterial biosensor system for arsenic detection holds great promise for making integrated measurement device. Bioeng. Bugs 2011, 2, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Duford, D.A.; Xi, Y.; Salin, E.D. Enzyme inhibition-based determination of pesticide residues in vegetable and soil in centrifugal microfluidic devices. Anal. Chem. 2013, 85, 7834–7841. [Google Scholar] [CrossRef] [PubMed]
- Foudeh, A.M.; Brassard, D.; Tabrizian, M.; Veres, T. Rapid and multiplex detection of Legionella’s RNA using digital microfluidics. Lab Chip 2015, 15, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Charles, P.T.; Adams, A.A.; Deschamps, J.R.; Veitch, S.; Hanson, A.; Kusterbeck, A.W. Detection of explosives in a dynamic marine environment using a moored TNT immunosensor. Sensors 2014, 14, 4074–4085. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Loke, W.K.; Tan, Y.T.; Nguyen, N.-T. A lab-on-a-chip for detection of nerve agent sarin in blood. Lab Chip 2008, 8, 885–891. [Google Scholar] [CrossRef] [PubMed]
- De Santis, R.; Ciammaruconi, A.; Faggioni, G.; Fillo, S.; Gentile, B.; Di Giannatale, E.; Ancora, M.; Lista, F. High throughput MLVA-16 typing for Brucella based on the microfluidics technology. BMC Microbiol. 2011, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dulay, S.B.; Gransee, R.; Julich, S.; Tomaso, H.; O’Sullivan, C.K. Automated microfluidically controlled electrochemical biosensor for the rapid and highly sensitive detection of Francisella tularensis. Biosens. Bioelectron. 2014, 59, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Matatagui, D.; Fontecha, J.L.; Fernández, M.J.; Gràcia, I.; Cané, C.; Santos, J.P.; Horrillo, M.C. Love-wave sensors combined with microfluidics for fast detection of biological warfare agents. Sensors 2014, 14, 12658–12669. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.S.; Haswell, S.J.; Lye, G.J.; Bracewell, D.G. Microfluidic chromatography for early stage evaluation of biopharmaceutical binding and separation conditions. Sep. Sci. Technol. 2011, 46, 185–194. [Google Scholar] [CrossRef]
- Chen, D.L.; Ismagilov, R.F. Microfluidic cartridges preloaded with nanoliter plugs of reagents: An alternative to 96-well plates for screening. Curr. Opin. Chem. Biol. 2006, 10, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xing, D. Miniaturized PCR chips for nucleic acid amplification and analysis: Latest advances and future trends. Nucleic Acids Res. 2007, 35, 4223–4237. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Mahalanabis, M.; Chang, J.; Carey, B.; Hsieh, C.; Stanley, A.; Odell, C.A.; Mitchell, P.; Feldman, J.; Pollock, N.R.; et al. Microfluidic chip for molecular amplification of influenza a RNA in human respiratory specimens. PLoS ONE 2012, 7, e33176. [Google Scholar] [CrossRef] [PubMed]
- Schell, W.A.; Benton, J.L.; Smith, P.B.; Poore, M.; Rouse, J.L.; Boles, D.J.; Johnson, M.D.; Alexander, B.D.; Pamula, V.K.; Eckhardt, A.E.; et al. Evaluation of a digital microfluidic real-time PCR platform to detect DNA of Candida albicans in blood. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2237–2245. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Segawa, T.; Okabe, S. Simultaneous quantification of multiple food- and waterborne pathogens by use of microfluidic quantitative PCR. Appl. Environ. Microbiol. 2013, 79, 2891–2898. [Google Scholar] [CrossRef] [PubMed]
- Spurgeon, S.L.; Jones, R.C.; Ramakrishnan, R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE 2008, 3, e1662. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhong, J.F. Microfluidic devices for high-throughput gene expression profiling of single hESC-derived neural stem cells. Methods Mol. Biol. 2008, 438, 293–303. [Google Scholar] [PubMed]
- May-Panloup, P.; Ferre-L’Hotellier, V.; Moriniere, C.; Marcaillou, C.; Lemerle, S.; Malinge, M.-C.; Coutolleau, A.; Lucas, N.; Reynier, P.; Descamps, P.; et al. Molecular characterization of corona radiata cells from patients with diminished ovarian reserve using microarray and microfluidic-based gene expression profiling. Hum. Reprod. 2012, 27, 829–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, K.J.; Hughes, E.M.; Dyer, C.E.; Greenman, J.; Haswell, S.J. Integrated RNA extraction and RT-PCR for semi-quantitative gene expression studies on a microfluidic device. Lab. Investig. 2013, 93, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Mellors, J.S.; Gorbounov, V.; Ramsey, R.S.; Ramsey, J.M. Fully integrated glass microfluidic device for performing high-efficiency capillary electrophoresis and electrospray ionization mass spectrometry. Anal. Chem. 2008, 80, 6881–6887. [Google Scholar] [CrossRef] [PubMed]
- Focke, M.; Mark, D.; Stumpf, F.; Müller, M.; Roth, G.; Zengerle, R.; von Stetten, F. Microfluidic cartridges for DNA purification and genotyping processed in standard laboratory instruments. In Proceedings of the SPIE, Smart Sensors, Actuators, and MEMS V, Prague, Czech Republic, 18–20 April 2011; Schmid, U., Sánchez-Rojas, J.L., Leester-Schaedel, M., Eds.; Volume 8066.
- Martinez, A.W.; Phillips, S.T.; Butte, M.J.; Whitesides, G.M. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. Int. Ed. 2007, 46, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M.; Carrilho, E. Diagnostics for the developing world: Microfluidic paper-based analytical devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R. When microfluidic devices go bad. Anal. Chem. 2005, 77, 429A–432A. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Khodakov, D.A.; Ellis, A.V.; Voelcker, N.H. Surface modification for PDMS-based microfluidic devices. Electrophoresis 2012, 33, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Barbulovic-Nad, I.; Wheeler, A.R. Cell Assays in Microfluidics. In Encyclopedia of Microfluidics and Nanofluidics; Springer: Berlin, Germany, 2008; pp. 209–216. [Google Scholar]
- Xiong, B.; Ren, K.; Shu, Y.; Chen, Y.; Shen, B.; Wu, H. Recent developments in microfluidics for cell studies. Adv. Mater. 2014, 26, 5525–5532. [Google Scholar] [CrossRef] [PubMed]
- Borenstein, J.T.; Vunjak-Novakovic, G. Engineering tissue with BioMEMS. IEEE Pulse 2011, 2, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Tourovskaia, A.; Fauver, M.; Kramer, G.; Simonson, S.; Neumann, T. Tissue-engineered microenvironment systems for modeling human vasculature. Exp. Biol. Med. 2014, 239, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Theberge, A.B.; Yu, J.; Young, E.W.K.; Ricke, W.A.; Bushman, W.; Beebe, D.J. Microfluidic multiculture assay to analyze biomolecular signaling in angiogenesis. Anal. Chem. 2015, 87, 3239–3246. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Duffy, S.P.; Matthews, K.; Santoso, A.T.; Scott, M.D.; Ma, H. Microfluidic analysis of red blood cell deformability. J. Biomech. 2014, 47, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Grosberg, A.; Alford, P.W.; McCain, M.L.; Parker, K.K. Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip. Lab Chip 2011, 11, 4165–4173. [Google Scholar] [CrossRef] [PubMed]
- Yeom, E.; Kang, Y.J.; Lee, S. Changes in velocity profile according to blood viscosity in a microchannel. Biomicrofluidics 2014, 8, 034110:1–034110:11. [Google Scholar] [CrossRef] [PubMed]
- Yeom, E.; Jun Kang, Y.; Joon Lee, S. Hybrid system for ex vivo hemorheological and hemodynamic analysis: A feasibility study. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tomaiuolo, G.; Lanotte, L.; D’Apolito, R.; Cassinese, A.; Guido, S. Microconfined flow behavior of red blood cells. Med. Eng. Phys. 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lv, X.; Ostrovidov, S.; Shi, X.; Zhang, N.; Liu, J. Biomimetic microfluidic device for in vitro antihypertensive drug evaluation. Mol. Pharm. 2014, 11, 2009–2015. [Google Scholar] [CrossRef] [PubMed]
- McCain, M.L.; Sheehy, S.P.; Grosberg, A.; Goss, J.A.; Parker, K.K. Recapitulating maladaptive, multiscale remodeling of failing myocardium on a chip. Proc. Natl. Acad. Sci. USA 2013, 110, 9770–9775. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, G.A.; Nguyen, M.-D.; Estrada, R.; Parichehreh, V.; Hamid, T.; Ismahil, M.A.; Prabhu, S.D.; Sethu, P. Microfluidic cardiac cell culture model (μCCCM). Anal. Chem. 2010, 82, 7581–7587. [Google Scholar] [CrossRef] [PubMed]
- Yasotharan, S.; Pinto, S.; Sled, J.G.; Bolz, S.-S.; Günther, A. Artery-on-a-chip platform for automated, multimodal assessment of cerebral blood vessel structure and function. Lab Chip 2015, 15, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Oh, S.; Lee, H.J.; Lee, J.Y.; Lee, H.K.; Jeon, N.L. Engineering a blood vessel network module for body-on-a-chip applications. J. Lab. Autom. 2015, 20, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Hattori, K.; Munehira, Y.; Kobayashi, H.; Satoh, T.; Sugiura, S.; Kanamori, T. Microfluidic perfusion culture chip providing different strengths of shear stress for analysis of vascular endothelial function. J. Biosci. Bioeng. 2014, 118, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Hald, E.S.; Steucke, K.E.; Reeves, J.A.; Win, Z.; Alford, P.W. Long-term vascular contractility assay using genipin-modified muscular thin films. Biofabrication 2014, 6, 045005:1–045005:11. [Google Scholar] [CrossRef] [PubMed]
- Dominical, V.M.; Vital, D.M.; O’Dowd, F.; Saad, S.T.O.; Costa, F.F.; Conran, N. In vitro microfluidic model for the study of vaso-occlusive processes. Exp. Hematol. 2015, 43, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.G.; Benipal, P.K.; Cheng, X.; Burdorf, L.; Azimzadeh, A.M.; Pierson, R.N. Four-dimensional characterization of thrombosis in a live-cell, shear-flow assay: Development and application to xenotransplantation. PLoS ONE 2015, 10, e0123015. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Li, F.; Lv, J.; He, Y.; Lu, D.; Yamada, T.; Ono, N. Microfluidic analysis of pressure drop and flow behavior in hypertensive micro vessels. Biomed. Microdevices 2015, 17, 1387–2176. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Bol, L.; Galas, J.-C.; Hillaireau, H.; Potier, I.L.; Nicolas, V.; Haghiri-Gosnet, A.-M.; Fattal, E.; Taverna, M. A microdevice for parallelized pulmonary permeability studies. Biomed. Microdevices 2014, 16, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-C.; Hsieh, M.-H.; Liu, C.-C.; Pan, H.-J.; Liao, W.-Y.; Cheng, J.-Y.; Kuo, P.-L.; Lee, C.-H. Modulating chemotaxis of lung cancer cells by using electric fields in a microfluidic device. Biomicrofluidics 2014, 8, 024107:1–024107:12. [Google Scholar] [CrossRef] [PubMed]
- Sellgren, K.L.; Butala, E.J.; Gilmour, B.P.; Randell, S.H.; Grego, S. A biomimetic multicellular model of the airways using primary human cells. Lab Chip 2014, 14, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Ling, T.-Y.; Liu, Y.-L.; Huang, Y.-K.; Gu, S.-Y.; Chen, H.-K.; Ho, C.-C.; Tsao, P.-N.; Tung, Y.-C.; Chen, H.-W.; Cheng, C.-H.; et al. Differentiation of lung stem/progenitor cells into alveolar pneumocytes and induction of angiogenesis within a 3D gelatin—Microbubble scaffold. Biomaterials 2014, 35, 5660–5669. [Google Scholar] [CrossRef] [PubMed]
- Punde, T.H.; Wu, W.-H.; Lien, P.-C.; Chang, Y.-L.; Kuo, P.-H.; Chang, M.D.-T.; Lee, K.-Y.; Huang, C.-D.; Kuo, H.-P.; Chan, Y.-F.; et al. A biologically inspired lung-on-a-chip device for the study of protein-induced lung inflammation. Integr. Biol. 2015, 7, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hillier, C.; Komenda, P.; Lobato de Faria, R.; Levin, D.; Zhang, M.; Lin, F. A microfluidic platform for evaluating neutrophil chemotaxis induced by sputum from COPD patients. PLoS ONE 2015, 10, e0126523. [Google Scholar] [CrossRef] [PubMed]
- Cortez-Jugo, C.; Qi, A.; Rajapaksa, A.; Friend, J.R.; Yeo, L.Y. Pulmonary monoclonal antibody delivery via a portable microfluidic nebulization platform. Biomicrofluidics 2015, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rochow, N.; Manan, A.; Wu, W.-I.; Fusch, G.; Monkman, S.; Leung, J.; Chan, E.; Nagpal, D.; Predescu, D.; Brash, J.; et al. An integrated array of microfluidic oxygenators as a neonatal lung assist device: In vitro characterization and in vivo demonstration. Artif. Organs 2014, 38, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Xu, Z.; Liu, F.; Wang, H.; Wen, J.; Shao, S.; Zhang, L.; Wang, L.; Liu, C.; Lu, J.; et al. Continual exposure to cigarette smoke extracts induces tumor-Like transformation of human nontumor bronchial epithelial cells in a microfluidic chip. J. Thorac. Oncol. 2014, 9, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Felder, M.; Stucki, A.O.; Stucki, J.D.; Geiser, T.; Guenat, O.T. The potential of microfluidic lung epithelial wounding: Towards in vivo-like alveolar microinjuries. Integr. Biol. 2014, 6, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadou, I.; Trabalza, A.; Ellison, S.M.; Gharun, K.; Mazarakis, N.D. Specific retrograde transduction of spinal motor neurons using lentiviral vectors targeted to presynaptic NMJ receptors. Mol. Ther. 2014, 22, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Campbell, G.E.; Szpankowski, L.; Goldstein, L.S.B.; Encalada, S.E. Characterizing the composition of molecular motors on moving axonal cargo using “cargo mapping” analysis. J. Vis. Exp. 2014, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Martin, S.; Papadopulos, A.; Harper, C.B.; Mavlyutov, T.A.; Niranjan, D.; Glass, N.R.; Cooper-White, J.J.; Sibarita, J.-B.; Choquet, D.; et al. Control of autophagosome axonal retrograde flux by presynaptic activity unveiled using botulinum neurotoxin type A. J. Neurosci. 2015, 35, 6179–6194. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhou, Y.; Weissmiller, A.M.; Pearn, M.L.; Mobley, W.C.; Wu, C. Real-time imaging of axonal transport of quantum dot-labeled BDNF in primary neurons. J. Vis. Exp. 2014, 91, e51899. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Bushell, T.J.; Zagnoni, M. Chemically induced synaptic activity between mixed primary hippocampal co-cultures in a microfluidic system. Integr. Biol. 2014, 6, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ferreira, M.M.; Heilshorn, S.C. Small-molecule axon-polarization studies enabled by a shear-free microfluidic gradient generator. Lab Chip 2014, 14, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Fillmore, H.L.; Vouri, M.; Pilkington, G.J. Brain tumor cell line authentication, an efficient alternative to capillary electrophoresis by using a microfluidics-based system. Neuro. Oncol. 2014, 16, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Pollen, A.A.; Nowakowski, T.J.; Shuga, J.; Wang, X.; Leyrat, A.A.; Lui, J.H.; Li, N.; Szpankowski, L.; Fowler, B.; Chen, P.; et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014, 32, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Kerman, B.E.; Kim, H.J.; Padmanabhan, K.; Mei, A.; Georges, S.; Joens, M.S.; Fitzpatrick, J.A.J.; Jappelli, R.; Chandross, K.J.; August, P.; et al. In vitro myelin formation using embryonic stem cells. Development 2015, 142, 2213–2225. [Google Scholar] [CrossRef] [PubMed]
- Nery, F.C.; da Hora, C.C.; Yaqub, U.; Zhang, X.; McCarthy, D.M.; Bhide, P.G.; Irimia, D.; Breakefield, X.O. New methods for investigation of neuronal migration in embryonic brain explants. J. Neurosci. Methods 2015, 239, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.-Y.; He, M.; Hou, Q.-Q.; Sheng, A.-L.; Yuan, L.; Liu, F.; Liu, W.-W.; Li, G.; Jiang, X.-Y.; Luo, Z.-G. Semaphorin 3A activates the guanosine triphosphatase Rab5 to promote growth cone collapse and organize callosal axon projections. Sci. Signal. 2014, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, L.; Tegenge, M.A.; Shrestha, S.; Ganesh Kumar, N.; Malik, A.; Mithal, A.; Hosmane, S.; Venkatesan, A. Toll-like receptor 4 deficiency impairs microglial phagocytosis of degenerating axons. Glia 2014, 62, 1982–1991. [Google Scholar] [CrossRef] [PubMed]
- Fournier, A.J.; Rajbhandari, L.; Shrestha, S.; Venkatesan, A.; Ramesh, K.T. In vitro and in situ visualization of cytoskeletal deformation under load: Traumatic axonal injury. FASEB J. 2014, 28, 5277–5287. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Sherrod, S.D.; Goodwin, C.R.; Brewer, B.; Yang, L.; Garbett, K.A.; Li, D.; McLean, J.A.; Wikswo, J.P.; Mirnics, K. Metabolic consequences of interleukin-6 challenge in developing neurons and astroglia. J. Neuroinflamm. 2014, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Kaplan, S.V.; Gehringer, R.C.; Limbocker, R.A.; Johnson, M.A. Localized drug application and sub-second voltammetric dopamine release measurements in a brain slice perfusion device. Anal. Chem. 2014, 86, 4151–4156. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, S.; Yu, X.; Liu, Z.; Wang, F.; Li, W.T.; Cheng, S.H.; Dai, Q.; Shi, P. High-throughput mapping of brain-wide activity in awake and drug-responsive vertebrates. Lab Chip 2015, 15, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Joshi, P.; Mastrangelo, R.; Francolini, M.; Verderio, C.; Matteoli, M. Testing Aβ toxicity on primary CNS cultures using drug-screening microfluidic chips. Lab Chip 2014, 14, 2860–2866. [Google Scholar] [CrossRef] [PubMed]
- Booth, R.; Kim, H. Permeability analysis of neuroactive drugs through a dynamic microfluidic in vitro blood–brain barrier model. Ann. Biomed. Eng. 2014, 42, 2379–2391. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Abdelfattah, A.S.; Zhao, Y.; Ruangkittisakul, A.; Ballanyi, K.; Campbell, R.E.; Harrison, D.J. Microfluidic cell sorter-aided directed evolution of a protein-based calcium ion indicator with an inverted fluorescent response. Integr. Biol. 2014, 6, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Coquinco, A.; Kojic, L.; Wen, W.; Wang, Y.T.; Jeon, N.L.; Milnerwood, A.J.; Cynader, M. A microfluidic based in vitro model of synaptic competition. Mol. Cell. Neurosci. 2014, 60, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Deleglise, B.; Magnifico, S.; Duplus, E.; Vaur, P.; Soubeyre, V.; Belle, M.; Vignes, M.; Viovy, J.-L.; Jacotot, E.; Peyrin, J.-M.; et al. B-amyloid induces a dying-back process and remote trans-synaptic alterations in a microfluidic-based reconstructed neuronal network. Acta Neuropathol. Commun. 2014, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xu, M.; Yang, M.; Yang, Y.; Li, Y.; Deng, J.; Ruan, L.; Liu, J.; Du, S.; Liu, X.; et al. An ALS-mutant TDP-43 neurotoxic peptide adopts an anti-parallel -structure and induces TDP-43 redistribution. Hum. Mol. Genet. 2014, 23, 6863–6877. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Mikheev, A.M.; Huynh, W.; Monnat, R.J.; Rostomily, R.C.; Folch, A. Parallel microfluidic chemosensitivity testing on individual slice cultures. Lab Chip 2014, 14, 4540–4551. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Zheng, W.; Xiao, L.; Zhang, W.; Jiang, X. Engineering a 3D vascular network in hydrogel for mimicking a nephron. Lab Chip 2013, 13, 1612–1618. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, R.; Alberto, G.; Legendre, A.; Paullier, P.; Naudot, M.; Fleury, M.J.; Jacques, S.; Griscom, L.; Leclerc, E. Investigation of expression and activity levels of primary rat hepatocyte detoxication genes under various flow rates and cell densities in microfluidic biochips. Biotechnol. Prog. 2014, 30, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Hung, P.J.; Lee, L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007, 97, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.N.; Green, B.J.; Altamentova, S.M.; Rocheleau, J.V. A microfluidic device designed to induce media flow throughout pancreatic islets while limiting shear-induced damage. Lab Chip 2013, 13, 4374–4384. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.; Kim, M.J.; Hwang, Y.H.; Jeon, E.A.; Kang, A.R.; Lee, S.-H.; Lee, D.Y. Microfluidics-generated pancreatic islet microfibers for enhanced immunoprotection. Biomaterials 2013, 34, 8122–8130. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Wang, Y.; Mendoza-Elias, J.E.; Adewola, A.F.; Harvat, T.A.; Kinzer, K.; Gutierrez, D.; Qi, M.; Eddington, D.T.; Oberholzer, J. Dual microfluidic perifusion networks for concurrent islet perifusion and optical imaging. Biomed. Microdevices 2012, 14, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Lee, P.J.; Hung, P.J.; Johnson, T.; Lee, L.P.; Mofrad, M.R.K. Microfluidic environment for high density hepatocyte culture. Biomed. Microdevices 2008, 10, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, R.; Griscom, L.; Prot, J.M.; Legallais, C.; Leclerc, E. Behavior of HepG2/C3A cell cultures in a microfluidic bioreactor. Biochem. Eng. J. 2011, 53, 172–181. [Google Scholar] [CrossRef]
- Illa, X.; Vila, S.; Yeste, J.; Peralta, C.; Gracia-Sancho, J.; Villa, R. A novel modular bioreactor to in vitro study the hepatic sinusoid. PLoS ONE 2014, 9, e111864. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Stevens, K.R.; Schwartz, R.E.; Alejandro, B.S.; Huang, J.H.; Bhatia, S.N. Micropatterned cell-cell interactions enable functional encapsulation of primary hepatocytes in hydrogel microtissues. Tissue Eng. Part A 2014, 20, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Domansky, K.; Inman, W.; Serdy, J.; Dash, A.; Lim, M.H.M.; Griffith, L.G. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip 2010, 10, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Suh, K.-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Li, D.; Gao, N.; Shi, Q.; Hou, H. Hepatogenic differentiation of mesenchymal stem cells using microfluidic chips. Biotechnol. J. 2008, 3, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-C.; Chang, Y.-J.; Chen, W.-C.; Harn, H.I.-C.; Tang, M.-J.; Wu, C.-C. Enhancement of renal epithelial cell functions through microfluidic-based coculture with adipose-derived stem cells. Tissue Eng. Part A 2013, 19, 2024–2034. [Google Scholar] [CrossRef] [PubMed]
- Ghazalli, N.; Mahdavi, A.; Feng, T.; Jin, L.; Kozlowski, M.T.; Hsu, J.; Riggs, A.D.; Tirrell, D.A.; Ku, H.T. Postnatal pancreas of mice contains tripotent progenitors capable of giving rise to duct, acinar, and endocrine cells in vitro. Stem Cells Dev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Ogunwobi, O.O.; Chen, T.; Zhang, J.; George, T.J.; Liu, C.; Fan, Z.H. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip 2014, 14, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Thege, F.I.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Kochman, M.L.; Hollingsworth, M.A.; Rhim, A.D.; Kirby, B.J. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: Characterization, optimization and downstream analysis. Lab Chip 2014, 14, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Thege, F.I.; Santana, S.M.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Maggs, L.R.; Kochman, M.L.; Ginsberg, G.G.; Lieb, J.G.; et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology 2014, 146, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ingber, D.E. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; Sung, J.H.; Yang, J.; Yu, C.; Yu, J.; March, J.C.; Shuler, M.L. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic “body-on-a-chip” devices. Biomed. Microdevices 2012, 14, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, J.W.; Choi, I.; Kim, Y.C.; Lee, J.B.; Sung, J.H. A microfluidic device with 3D hydrogel villi scaffold to simulate intestinal absorption. J. Nanosci. Nanotechnol. 2013, 13, 7220–7228. [Google Scholar] [CrossRef] [PubMed]
- Legendre, A.; Baudoin, R.; Alberto, G.; Paullier, P.; Naudot, M.; Bricks, T.; Brocheton, J.; Jacques, S.; Cotton, J.; Leclerc, E. Metabolic characterization of primary rat hepatocytes cultivated in parallel microfluidic biochips. J. Pharm. Sci. 2013, 102, 3264–3276. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Yamamoto, T.; Sakai, H.; Sakai, Y.; Fujii, T. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 2008, 8, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Pasirayi, G.; Scott, S.M.; Islam, M.; O’Hare, L.; Bateson, S.; Ali, Z. Low cost microfluidic cell culture array using normally closed valves for cytotoxicity assay. Talanta 2014, 129, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Emami, K.; Wu, H.; Sun, W. Biofabrication of a three-dimensional liver micro-organ as an in vitro drug metabolism model. Biofabrication 2010, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.C.; Emami, K.; Jeevarajan, A.; Wu, H.; Sun, W. Microprinting of liver micro-organ for drug metabolism study. Methods Mol. Biol. 2011, 671, 219–238. [Google Scholar] [PubMed]
- Ramadan, Q.; Jafarpoorchekab, H.; Huang, C.; Silacci, P.; Carrara, S.; Koklü, G.; Ghaye, J.; Ramsden, J.; Ruffert, C.; Vergeres, G.; et al. NutriChip: Nutrition analysis meets microfluidics. Lab Chip 2013, 13, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Snouber, L.C.; Bunescu, A.; Naudot, M.; Legallais, C.; Brochot, C.; Dumas, M.E.; Elena-Herrmann, B.; Leclerc, E. Metabolomics-on-a-chip of hepatotoxicity induced by anticancer drug flutamide and its active metabolite hydroxyflutamide using HepG2/C3a microfluidic biochips. Toxicol. Sci. 2013, 132, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Dhumpa, R.; Truong, T.M.; Wang, X.; Roper, M.G. Measurement of the entrainment window of islets of Langerhans by microfluidic delivery of a chirped glucose waveform. Integr. Biol. 2015, 7, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Lomasney, A.R.; Yi, L.; Roper, M.G. Simultaneous monitoring of insulin and islet amyloid polypeptide secretion from islets of langerhans on a microfluidic device. Anal. Chem. 2013, 85, 7919–7925. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.F.; Wang, Y.; Blake, A.; Yu, G.; Harvat, T.A.; Jeon, H.; Oberholzer, J.; Eddington, D.T. Islet preconditioning via multimodal microfluidic modulation of intermittent hypoxia. Anal. Chem. 2012, 84, 1987–1993. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, N.; Ricci, K.B.; Groszek, J.; Marmerstein, J.T.; Fissell, W.H. Albumin handling by renal tubular epithelial cells in a microfluidic bioreactor. Biotechnol. Bioeng. 2012, 109, 797–803. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, G.J.; Chang, J.Y.; Glahn, R.P.; Shuler, M.L. Development of a gastrointestinal tract microscale cell culture analog to predict drug transport. Mol. Celullar Biomech. 2008, 5, 119–132. [Google Scholar]
- Nourmohammadzadeh, M.; Lo, J.F.; Bochenek, M.; Mendoza-Elias, J.E.; Wang, Q.; Li, Z.; Zeng, L.; Qi, M.; Eddington, D.T.; Oberholzer, J.; et al. Microfluidic array with integrated oxygenation control for real-time live-cell imaging: Effect of hypoxia on physiology of microencapsulated pancreatic islets. Anal. Chem. 2013, 85, 11240–11249. [Google Scholar] [CrossRef] [PubMed]
- Bricks, T.; Paullier, P.; Legendre, A.; Fleury, M.J.; Zeller, P.; Merlier, F.; Anton, P.M.; Leclerc, E. Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines. Toxicol. Vitr. 2014, 28, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Ikeda, T.; Nakayama, H.; Sakai, Y.; Fujii, T. An on-chip small intestine-liver model for pharmacokinetic studies. J. Lab. Autom. 2015, 20, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Matharu, Z.; Patel, D.; Gao, Y.; Haque, A.; Zhou, Q.; Revzin, A. Detecting transforming growth factor-β release from liver cells using an aptasensor integrated with microfluidics. Anal. Chem. 2014, 86, 8865–8872. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, Y.; Sonkusale, S.R. Microfluidic optoelectronic sensor for salivary diagnostics of stomach cancer. Biosens. Bioelectron. 2015, 67, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Murphy, R.G.; Seibel, E.J. Beyond isolated cells: Microfluidic transport of large tissue for pancreatic cancer diagnosis. Proc. SPIE 2015, 9320, 1–29. [Google Scholar]
- Leonard, E.F. Technical approaches toward ambulatory ESRD therapy. Seminars Dial. 2009, 22, 658–660. [Google Scholar] [CrossRef] [PubMed]
- Leonard, E.F.; Cortell, S.; Jones, J. The path to wearable ultrafiltration and dialysis devices. Blood Purification 2011, 31, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Kim, H.J.; Fraser, J.P.; Shea, D.E.; Khan, M.; Bahinski, A.; Hamilton, G.A.; Ingber, D.E. Microfabrication of human organs-on-chips. Nat. Protoc. 2013, 8, 2135–2157. [Google Scholar] [CrossRef] [PubMed]
- Legendre, A.; Jacques, S.; Dumont, F.; Cotton, J.; Paullier, P.; Fleury, M.J.; Leclerc, E. Investigation of the hepatotoxicity of flutamide: Pro-survival/apoptotic and necrotic switch in primary rat hepatocytes characterized by metabolic and transcriptomic profiles in microfluidic liver biochips. Toxicol. Vitr. 2014, 28, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Hamon, J.; Claude, I.; Jellali, R.; Naudot, M.; Bois, F. Investigation of acetaminophen toxicity in HepG2/C3a microscale cultures using a system biology model of glutathione depletion. Cell Biol. Toxicol. 2015, 31, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Snouber, L.C.; Jacques, S.; Monge, M.; Legallais, C.; Leclerc, E. Transcriptomic analysis of the effect of ifosfamide on MDCK cells cultivated in microfluidic biochips. Genomics 2012, 100, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Snouber, L.C.; Letourneur, F.; Chafey, P.; Broussard, C.; Monge, M.; Legallais, C.; Leclerc, E. Analysis of transcriptomic and proteomic profiles demonstrates improved Madin-Darby canine kidney cell function in a renal microfluidic biochip. Biotechnol. Prog. 2011, 28, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Snouber, L.C.; Aninat, C.; Grsicom, L.; Madalinski, G.; Brochot, C.; Poleni, P.E.; Razan, F.; Guillouzo, C.G.; Legallais, C.; Corlu, A.; et al. Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnol. Bioeng. 2013, 110, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Mahler, G.J.; Esch, M.B.; Glahn, R.P.; Shuler, M.L. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009, 104, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Hamon, J.; Legendre, A.; Bois, F.Y. Integration of pharmacokinetic and NRF2 system biology models to describe reactive oxygen species production and subsequent glutathione depletion in liver microfluidic biochips after flutamide exposure. Toxicol. Vitr. 2014, 28, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.D.; Berry, S.M.; Powers, G.L.; Beebe, D.J.; Alarid, E.T. Hormonally responsive breast cancer cells in a microfluidic co-culture model as a sensor of microenvironmental activity. Integr. Biol. 2013, 5, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Hannanta-anan, P.; Chau, M.; Kim, Y.S.; Swartz, M.A.; Wu, M. Cooperative roles of SDF-1α and EGF gradients on tumor cell migration revealed by a robust 3D microfluidic model. PLoS ONE 2013, 8, e68422. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Wu, T.-L.; Huang, H.-R.; Li, C.-J.; Fu, H.-T.; Soong, Y.-K.; Lee, M.-Y.; Yao, D.-J. Isolation of motile spermatozoa with a microfluidic chip having a surface-modified microchannel. J. Lab. Autom. 2014, 19, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.; Hu, L.; Fiore, A.G.; Ardon, F.; Hickman, D.G.; Gilbert, R.O.; Suarez, S.S.; Wu, M. Microgrooves and fluid flows provide preferential passageways for sperm over pathogen Tritrichomonas foetus. Proc. Natl. Acad. Sci. USA 2015, 112, 5431–5436. [Google Scholar] [CrossRef] [PubMed]
- Ferrie, A.M.; Wang, C.; Deng, H.; Fang, Y. A label-free optical biosensor with microfluidics identifies an intracellular signalling wave mediated through the β2-adrenergic receptor. Integr. Biol. 2013, 5, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Ges, I.A.; Brindley, R.L.; Currie, K.P.M.; Baudenbacher, F.J. A microfluidic platform for chemical stimulation and real time analysis of catecholamine secretion from neuroendocrine cells. Lab Chip 2013, 13, 4663–4673. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.M.; Dong, T. Detection of stress hormones by a microfluidic-integrated polycarbazole/fullerene photodetector. In Proceedings of the 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013.
- Broccardo, C.J.; Schauer, K.L.; Kohrt, W.M.; Schwartz, R.S.; Murphy, J.P.; Prenni, J.E. Multiplexed analysis of steroid hormones in human serum using novel microflow tile technology and LC-MS/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 934, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Abdulwahab, S.; Choi, K.; Lafrenière, N.M.; Mudrik, J.M.; Gomaa, H.; Ahmado, H.; Behan, L.-A.; Casper, R.F.; Wheeler, A.R. A microfluidic technique for quantification of steroids in core needle biopsies. Anal. Chem. 2015, 87, 4688–4695. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.; Yndart, A.; Jayant, R.D.; Sagar, V.; Atluri, V.; Bhansali, S.; Nair, M. Electrochemical sensing method for point-of-care cortisol detection in human immunodeficiency virus-infected patients. Int. J. Nanomedicine 2015, 10, 677–685. [Google Scholar] [PubMed]
- Selimovic, A.; Erkal, J.L.; Spence, D.M.; Martin, R.S. Microfluidic device with tunable post arrays and integrated electrodes for studying cellular release. Analyst 2014, 139, 5686–5694. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, M.H.; Choi, K.; Ng, A.H.C.; Wheeler, A.R. A digital microfluidic electrochemical immunoassay. Lab Chip 2014, 14, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Arends, F.; Sellner, S.; Seifert, P.; Gerland, U.; Rehberg, M.; Lieleg, O. A microfluidics approach to study the accumulation of molecules at basal lamina interfaces. Lab Chip 2015, 15, 3326–3334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Smith, J.; Yarmush, M.L.; Nahmias, Y.; Kirby, B.J.; Murthy, S.K. Microfluidic enrichment of mouse epidermal stem cells and validation of stem cell proliferation in vitro. Tissue Eng. Part C Methods 2013, 19, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Jean, L.; Yang, L.; Majumdar, D.; Gao, Y.; Shi, M.; Brewer, B.M.; Li, D.; Webb, D.J. The Rho family GEF Asef2 regulates cell migration in three dimensional (3D) collagen matrices through myosin II. Cell Adh. Migr. 2014, 8, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Smith, K.; Kaya, T. Implementation of a microfluidic conductivity sensor—A potential sweat electrolyte sensing system for dehydration detection. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2014. [Google Scholar] [CrossRef]
- Rose, D.P.; Ratterman, M.E.; Griffin, D.K.; Hou, L.; Kelley-Loughnane, N.; Naik, R.R.; Hagen, J.A.; Papautsky, I.; Heikenfeld, J. Adhesive RFID sensor patch for monitoring of sweat electrolytes. IEEE Trans. Biomed. Eng. 2015, 62, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, S.; Huang, R.; Huang, Z.; Hu, B.; Zheng, W.; Yang, G.; Jiang, X. Evaluation of the effect of the structure of bacterial cellulose on full thickness skin wound repair on a microfluidic chip. Biomacromolecules 2015, 16, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.F.; Brennan, M.; Merchant, Z.; Chen, L.; Guo, S.; Eddington, D.T.; DiPietro, L.A. Microfluidic wound bandage: Localized oxygen modulation of collagen maturation. Wound Repair Regen. 2013, 21, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Berthier, E.; Young, E.W.K.; Beebe, D. Engineers are from PDMS-land, biologists are from Polystyrenia. Lab Chip 2012, 12, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Jiang, B.; Wang, D.; Zhang, W.; Wang, Z.; Jiang, X. A microfluidic flow-stretch chip for investigating blood vessel biomechanics. Lab Chip 2012, 12, 3441–3450. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Jiang, B.; Hao, Y.; Zhao, Y.; Zhang, W.; Jiang, X. Screening reactive oxygen species scavenging properties of platinum nanoparticles on a microfluidic chip. Biofabrication 2014, 6, 045004:1–045004:11. [Google Scholar] [CrossRef] [PubMed]
- Nalayanda, D.D.; Wang, Q.; Fulton, W.B.; Wang, T.-H.; Abdullah, F. Engineering an artificial alveolar-capillary membrane: A novel continuously perfused model within microchannels. J. Pediatr. Surg. 2010, 45, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Nalayanda, D.D.; Puleo, C.M.; Fulton, W.B.; Wang, T.-H.; Abdullah, F. Characterization of pulmonary cell growth parameters in a continuous perfusion microfluidic environment. Exp. Lung Res. 2007, 33, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.E.; Niles, J.A.; Vega, S.P.; Argueta, L.B.; Eastaway, A.; Cortiella, J. Modeling the lung: Design and development of tissue engineered macro- and micro-physiologic lung models for research use. Exp. Biol. Med. 2014, 239, 1135–1169. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Qiu, L.; Yang, X.; Zhou, Y.; Du, J.; Wang, H.; Sun, J.; Yang, C.; Jiang, J. Isolation of lung multipotent stem cells using a novel microfluidic magnetic activated cell sorting system. Cell Biol. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hoganson, D.M.; Pryor, H.I., II; Bassett, E.K.; Spool, I.D.; Vacanti, J.P. Lung assist device technology with physiologic blood flow developed on a tissue engineered scaffold platform. Lab Chip 2011, 11, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Kniazeva, T.; Hsiao, J.C.; Charest, J.L.; Borenstein, J.T. A microfluidic respiratory assist device with high gas permeance for artificial lung applications. Biomed. Microdevices 2011, 13, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Kniazeva, T.; Epshteyn, A.A.; Hsiao, J.C.; Kim, E.S.; Kolachalama, V.B.; Charest, J.L.; Borenstein, J.T. Performance and scaling effects in a multilayer microfluidic extracorporeal lung oxygenation device. Lab Chip 2012, 12, 1686–1695. [Google Scholar] [CrossRef] [PubMed]
- Kovach, K.M.; LaBarbera, M.A.; Moyer, M.C.; Cmolik, B.L.; van Lunteren, E.; Sen Gupta, A.; Capadona, J.R.; Potkay, J.A. In vitro evaluation and in vivo demonstration of a biomimetic, hemocompatible, microfluidic artificial lung. Lab Chip 2015, 15, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.M.; Blurton-Jones, M.; Rhee, S.W.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2005, 2, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Booth, R.; Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Shintu, L.; Baudoin, R.; Navratil, V.; Prot, J.-M.; Pontoizeau, C.; Defernez, M.; Blaise, B.J.; Domange, C.; Péry, A.R.; Toulhoat, P.; et al. Metabolomics-on-a-chip and predictive systems toxicology in microfluidic bioartificial organs. Anal. Chem. 2012, 84, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, R.; Legendre, A.; Jacques, S.; Cotton, J.; Bois, F.; Leclerc, E. Evaluation of a liver microfluidic biochip to predict in vivo clearances of seven drugs in rats. J. Pharm. Sci. 2014, 103, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Van Midwoud, P.M.; Verpoorte, E.; Groothuis, G.M.M. Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr. Biol. 2011, 3, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; El-Khatib, F.H.; Sinha, M.; Magyar, K.L.; McKeon, K.; Goergen, L.G.; Balliro, C.; Hillard, M.A.; Nathan, D.M.; Damiano, E.R. Outpatient glycemic control with a bionic pancreas in type 1 diabetes. N. Engl. J. Med. 2014, 371, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, Y.; Jia, L.; Mathewson, K.E.; Jang, K.-I.; Kim, J.; Fu, H.; Huang, X.; Chava, P.; Wang, R.; et al. Soft microfluidic assemblies of sensors, circuits, and radios for the skin. Science 2014, 344, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Sonner, Z.; Wilder, E.; Heikenfeld, J.; Kasting, G.; Beyette, F.; Swaile, D.; Sherman, F.; Joyce, J.; Hagen, J.; Kelley-Loughnane, N.; et al. The microfluidics of the eccrine sweat gland, including biomarker partitioning, transport, and biosensing implications. Biomicrofluidics 2015. [Google Scholar] [CrossRef] [PubMed]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012, 11, 191–200. [Google Scholar] [PubMed]
- Esch, M.B.; Mahler, G.J.; Stokol, T.; Shuler, M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 2014, 14, 3081–3092. [Google Scholar] [CrossRef] [PubMed]
- Shuler, M.L. Modeling life. Ann. Biomed. Eng. 2012, 40, 1399–1407. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Kato, K.; Kobayashi, M.; Kosaka, N.; Ochiya, T.; Ichiki, T. On-Chip Immunoelectrophoresis of Extracellular Vesicles Released from Human Breast Cancer Cells. PLoS ONE 2015, 10, e0123603. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.-T.; Chen, W.; Oh, B.-R.; Cornell, T.T.; Shanley, T.P.; Fu, J.; Kurabayashi, K. An integrated microfluidic platform for in situ cellular cytokine secretion immunophenotyping. Lab Chip 2012, 12, 4093–4101. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; King, T.L.; Shuler, M.L. The role of body-on-a-chip devices in drug and toxicity studies. Annu. Rev. Biomed. Eng. 2011, 13, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Vunjak-Novakovic, G.; Bhatia, S.; Chen, C.; Hirschi, K. HeLiVa platform: Integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res. Ther. 2013. [Google Scholar] [CrossRef] [PubMed]
- Prot, J.M.; Maciel, L.; Bricks, T.; Merlier, F.; Cotton, J.; Paullier, P.; Bois, F.Y.; Leclerc, E. First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol. Bioeng. 2014, 111, 2027–2040. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Hasenberg, T.; Jaenicke, A.; Lindner, M.; Lorenz, A.K.; Zech, J.; Garbe, L.-A.; Sonntag, F.; Hayden, P.; Ayehunie, S.; et al. Chip-based human liver–intestine and liver–skin co-cultures—A first step toward systemic repeated dose substance testing in vitro. Eur. J. Pharm. Biopharm. 2015. [Google Scholar] [CrossRef] [PubMed]
- Frey, O.; Misun, P.M.; Fluri, D.A.; Hengstler, J.G.; Hierlemann, A. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat. Commun. 2014, 5, 1–11. [Google Scholar]
- Kim, J.-Y.; Fluri, D.A.; Kelm, J.M.; Hierlemann, A.; Frey, O. 96-Well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. J. Lab. Autom. 2015, 20, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Fluri, D.A.; Marchan, R.; Boonen, K.; Mohanty, S.; Singh, P.; Hammad, S.; Landuyt, B.; Hengstler, J.G.; Kelm, J.M.; et al. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J. Biotechnol. 2015, 205, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Kam, C.; Shuler, M.L. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip 2010, 10, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Srinivasan, B.; Esch, M.B.; McLamb, W.T.; Bernabini, C.; Shuler, M.L.; Hickman, J.J. Using physiologically-based pharmacokinetic-guided “body-on-a-chip” systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med. 2014, 239, 1225–1239. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P.; Curtis, E.L.; Eagleton, Z.E.; Evans, B.C.; Kole, A.; Hofmeister, L.H.; Matloff, W.J. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013, 13, 3496–3511. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Dance, A. Correction for Dance, News Feature: Building benchtop human models. Proc. Natl. Acad. Sci. USA 2015, 112, 6773–6775. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Allen, S.G.; Ingram, P.N.; Buckanovich, R.; Merajver, S.D.; Yoon, E. Single-cell migration chip for chemotaxis-based microfluidic selection of heterogeneous cell populations. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, W.; Ryu, H.; Jeon, N.L. A microfluidic platform for quantitative analysis of cancer angiogenesis and intravasationa. Biomicrofluidics 2014. [Google Scholar] [CrossRef] [PubMed]
- Mattei, F.; Schiavoni, G.; De Ninno, A.; Lucarini, V.; Sestili, P.; Sistigu, A.; Fragale, A.; Sanchez, M.; Spada, M.; Gerardino, A.; et al. A multidisciplinary study using in vivo tumor models and microfluidic cell-on-chip approach to explore the cross-talk between cancer and immune cells. J. Immunotoxicol. 2014, 11, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, L.; Qin, L. High-throughput 3D cell invasion chip enables accurate cancer metastatic assays. J. Am. Chem. Soc. 2014, 136, 15257–15262. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Yue, W.; Yu, W.-K.; Liu, D.; Fong, C.-C.; Zhao, J.; Yang, M. Microfluidic platform for studying chemotaxis of adhesive cells revealed a gradient-dependent migration and acceleration of cancer stem cells. Anal. Chem. 2015, 87, 7098–7108. [Google Scholar] [CrossRef] [PubMed]
- Riahi, R.; Yang, Y.L.; Kim, H.; Jiang, L.; Wong, P.K.; Zohar, Y. A microfluidic model for organ-specific extravasation of circulating tumor cells. Biomicrofluidics 2014, 8, 024103. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Pei, Y.; Xie, M.; Jin, Z.-H.; Xiao, Y.-S.; Wang, Y.; Zhang, L.-N.; Li, Y.; Huang, W.-H. An artificial blood vessel implanted three-dimensional microsystem for modeling transvascular migration of tumor cells. Lab Chip 2015, 15, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Casavant, B.P.; Strotman, L.N.; Tokar, J.J.; Thiede, S.M.; Traynor, A.M.; Ferguson, J.S.; Lang, J.M.; Beebe, D. Paired diagnostic and pharmacodynamic analysis of rare non-small cell lung cancer cells enabled by the VerIFAST platform. Lab Chip 2014, 14, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-M.; Kim, M.S.; Moon, H.-S.; Yoo, C.E.; Park, D.; Kim, Y.J.; Han, K.-Y.; Lee, J.-Y.; Oh, J.H.; Kim, S.S.; et al. Fully automated circulating tumor cell isolation platform with large-volume capacity based on lab-on-a-disc. Anal. Chem. 2014, 86, 3735–3742. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Serizawa, M.; Sawada, T.; Takeda, K.; Takahashi, T.; Yamamoto, N.; Koizumi, F.; Koh, Y. A novel flow cytometry-based cell capture platform for the detection, capture and molecular characterization of rare tumor cells in blood. J. Transl. Med. 2014, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.F.; Yu, Y.H.; Chen, L.Y.; Fan, S.K.; Chou, H.Y.E.; Yang, J.T. A portable microfluidic device for the rapid diagnosis of cancer metastatic potential which is programmable for temperature and CO2. Lab Chip 2014, 14, 3621–3628. [Google Scholar] [CrossRef] [PubMed]
- Galletti, G.; Sung, M.S.; Vahdat, L.T.; Shah, M.A.; Santana, S.M.; Altavilla, G.; Kirby, B.J.; Giannakakou, P. Isolation of breast cancer and gastric cancer circulating tumor cells by use of an anti HER2-based microfluidic device. Lab Chip 2014, 14, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-T.; Zhao, L.; Shen, Q.; Garcia, M.A.; Wu, D.; Hou, S.; Song, M.; Xu, X.; OuYang, W.-H.; OuYang, W.W.-L.; et al. NanoVelcro chip for CTC enumeration in prostate cancer patients. Methods 2013, 64, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-Y.; Liu, H.-C.; Yen, L.-C.; Chang, J.-Y.; Huang, J.-J.; Wang, J.-Y.; Lin, S.-R. Decreasing relapse in colorectal cancer patients treated with cetuximab by using the activating KRAS detection chip. Tumor Biol. 2014, 35, 9639–9647. [Google Scholar] [CrossRef] [PubMed]
- Xue, P.; Wu, Y.; Guo, J.; Kang, Y. Highly efficient capture and harvest of circulating tumor cells on a microfluidic chip integrated with herringbone and micropost arrays. Biomed. Microdevices 2015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shiratsuchi, H.; Lin, J.; Chen, G.; Reddy, R.M.; Azizi, E.; Fouladdel, S.; Chang, A.C.; Lin, L.; Jiang, H.; et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014, 5, 12383–12397. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Jia, C.-P.; Jun-Yang; Sun, W.-J.; Wang, W.-T.; Zhang, H.-L.; Cong, H.; Jing, F.-X.; Mao, H.-J.; Jin, Q.-H.; et al. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens. Bioelectron. 2014, 51, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Zhu, Z.; Xu, Z.; He, T.; Li, E.; Guo, Z.; Liu, F.; Jiang, C.; Wang, Q. Cancer associated fibroblast-derived hepatocyte growth factor inhibits the paclitaxel-induced apoptosis of lung cancer A549 cells by up-regulating the PI3K/Akt and GRP78 signaling on a microfluidic platform. PLoS ONE 2015, 10, e0129593. [Google Scholar] [CrossRef] [PubMed]
- Ruppen, J.; Wildhaber, F.D.; Strub, C.; Hall, S.R.R.; Schmid, R.A.; Geiser, T.; Guenat, O.T. Towards personalized medicine: Chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip 2015, 15, 3076–3085. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Zervantonakis, I.K.; Chung, S.; Kamm, R.D.; Charest, J.L. In vitro model of tumor cell extravasation. PLoS ONE 2013, 8, e56910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bersini, S.; Jeon, J.S.; Dubini, G.; Arrigoni, C.; Chung, S.; Charest, J.L.; Moretti, M.; Kamm, R.D. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 2014, 35, 2454–2461. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Bersini, S.; Gilardi, M.; Dubini, G.; Charest, J.L.; Moretti, M.; Kamm, R.D. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. USA 2015, 112, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P.; Block, F.E.; Cliffel, D.E.; Goodwin, C.R.; Marasco, C.C.; Markov, D.A.; McLean, D.L.; McLean, J.A.; McKenzie, J.R.; Reiserer, R.S.; et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans. Biomed. Eng. 2013, 60, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W.W.M. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perestrelo, A.R.; Águas, A.C.P.; Rainer, A.; Forte, G. Microfluidic Organ/Body-on-a-Chip Devices at the Convergence of Biology and Microengineering. Sensors 2015, 15, 31142-31170. https://doi.org/10.3390/s151229848
Perestrelo AR, Águas ACP, Rainer A, Forte G. Microfluidic Organ/Body-on-a-Chip Devices at the Convergence of Biology and Microengineering. Sensors. 2015; 15(12):31142-31170. https://doi.org/10.3390/s151229848
Chicago/Turabian StylePerestrelo, Ana Rubina, Ana C. P. Águas, Alberto Rainer, and Giancarlo Forte. 2015. "Microfluidic Organ/Body-on-a-Chip Devices at the Convergence of Biology and Microengineering" Sensors 15, no. 12: 31142-31170. https://doi.org/10.3390/s151229848







