DNA Markers and FCSS Analyses Shed Light on the Genetic Diversity and Reproductive Strategy of Jatropha curcas L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
| Accession Id. | Geographical origin | Variety | Seeds | Plants |
|---|---|---|---|---|
| JcCLO | America/Mexico | Cruz de Loredo1 | 100 | 4 |
| JcHDE | America/Mexico | Playon de Mismaloya1 | 10 | 5 |
| JcBRA | America/Brazil | n.s.2 | 30 | 5 |
| JcSAM | America/Brazil | n.s.2 | 10 | 1 |
| JcLOL | America/South America | Lola | 200 | 20 |
| JcTOG | Africa/Togo | n.s.2 | 29 | 7 |
| JcENG | Africa/Tanzania | n.s.2 | 150 | 27 |
| JcBER | Africa/Western Africa | Berenice | 200 | 22 |
| JcESM | Africa/Western Africa | Esmeralda | 200 | 18 |
| JcKAR | Africa/Western Africa | Karima | 200 | 10 |
| JcSLK | Asia/Sri Lanka | Anil | 230 | 20 |
| JcPET | Asia/South-East Asia | Petra | 200 | 7 |
| JcSEL | Asia/South-East Asia | Selene | 200 | 19 |
| JcJOR3 | Asia/Jordan | n.s.2 | 100 | 5 |
| JcCHH | Asia/India (Chhattisgarh) | Chhattisgarh | 200 | 10 |
| JcUDA | Asia/India (Radjesthan) | Udaipur | 60 | 5 |
| JcKAK | Asia/India (Andhra Pradesh) | Kakinada | 200 | 4 |
| JcTUT | Asia/India (Tamilnadu) | Tuticorin | 60 | 5 |
| JcCOI | Asia/India (Tamilnadu ) | Coimbatore | 200 | 10 |
| Overall | 2,579 | 204 |
2.2. Ploidy Estimation and Flow Cytometric Seed Screen (FCSS) Analysis of Reproductive Modes
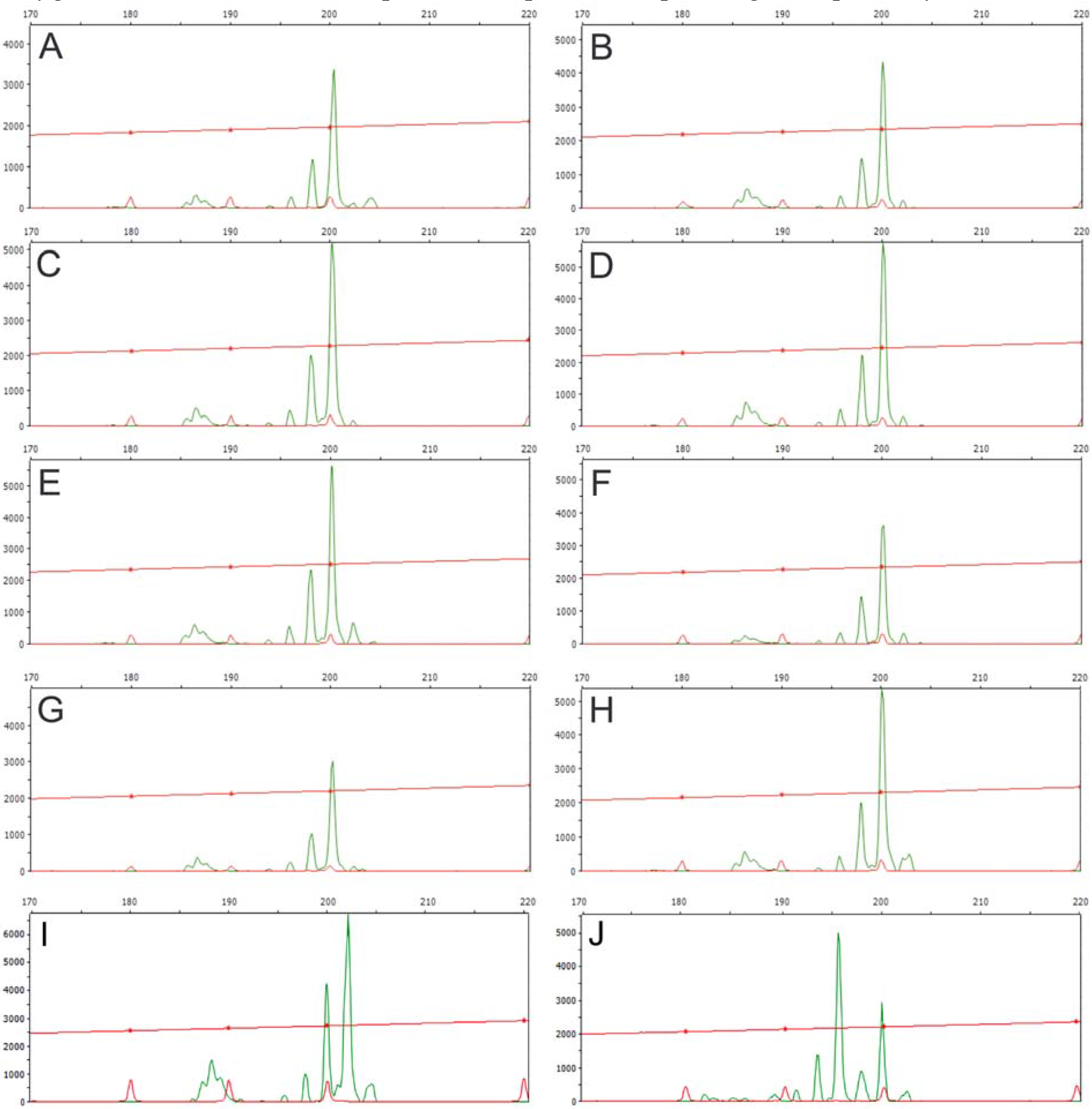
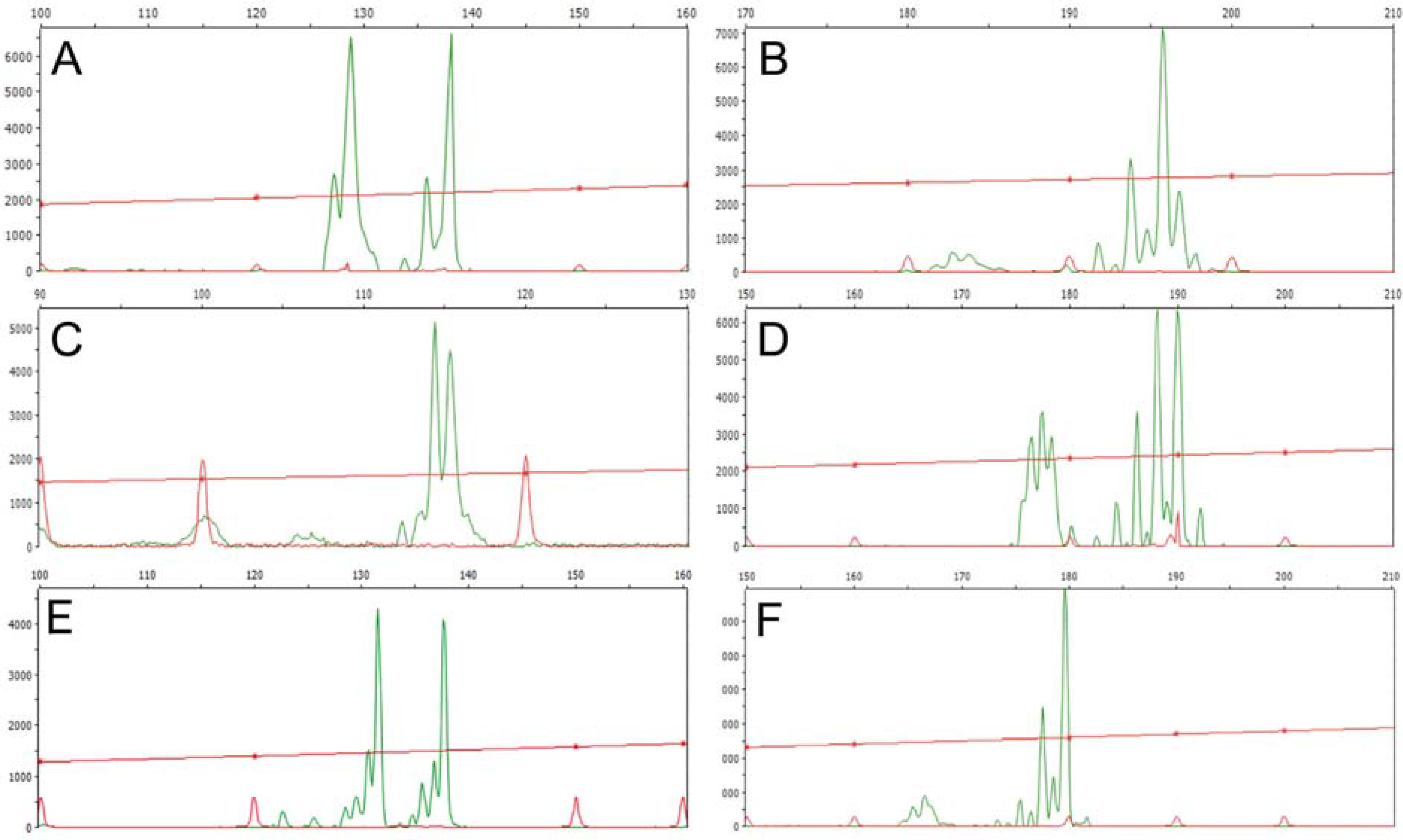
2.3. Genomic DNA Isolation and DNA Marker Detection
| Primer name | Sequence (5′-3′) | Basic information | ||
|---|---|---|---|---|
| RAPD | 10-mer primer | Ta (°C) | No. amplicons | Size range (bp) |
| OP-A1 | CAGGCCCTTC | 36 | 14 | 300–2,000 |
| OP-A13 | CAGCACCCAC | 36 | 16 | 100–1,500 |
| OP-E20 | AACGGTGACC | 36 | 9 | 300–2,000 |
| OP-G14 | GGATGAGACC | 36 | 8 | 200–2,100 |
| OP-M3 | GGGGGATGAG | 36 | 16 | 250–1,500 |
| OP-M13 | GGTGGTCAAC | 36 | 11 | 200–1,700 |
| OP-P2 | TCGGCACGAC | 36 | 16 | 170–2,500 |
| OP-P6 | GTGGGCTGAC | 36 | 13 | 200–2,000 |
| OP-Q2 | TCTGTCGGTC | 36 | 18 | 220–2,000 |
| OP-R8 | CCCGTTGCCT | 36 | 11 | 200–1,650 |
| Inter-SSR | SSR-anchored primer | Ta (°C) | No. amplicons | Size range (bp) |
| I-4 | (CA)8AC | 58 | 9 | 300–1,200 |
| I-15 | GTGC(AC)7 | 60 | 12 | 450–3,000 |
| I-19 | CCTGC(AC)7 | 61 | 10 | 400–2,000 |
| I-30 | (GT)6TG | 49 | 9 | 450–2,000 |
| I-32 | (AGC)4C | 59 | 6 | 400–2,100 |
| I-35 | (AGC)4GA | 59 | 10 | 480–2,800 |
| SSR | Forward and Reverse primers | Ta (°C) | Repeat | Expected size (bp) |
| JcSSR1 | GCAAGAGCTCCACAGTAGAAGA | 55 | (AG)15 | 168–187 |
| CTACCAACTCTTTCCAGTCC | ||||
| JcSSR2 | GCGCTAGAGATGCTGTTTTCT | 54 | (CT)13 | 123–138 |
| CACCACTCTGCAAATAGAAC | ||||
| JcSSR3 | GACAATGTTAATCGAAGGAG | 55 | (GA)24 | 164–195 |
| GACGGGAAAATATATGGCTA | ||||
| JcSSR4 | GTTTTCAGGATTGAATGCTCT | 55 | (AG)20 | 178–198 |
| CGCAATCCCATTTTTATTAT | ||||
| JcSSR5 | GATGGGCTCTTTGTAGCTTTT | 56 | (CT)8 | 90 |
| TTCATGCGACTAGAGCCTAC | ||||
| JcSSR6 | GTCTATTGAGTGGTTGCATTG | 55 | (TG)23 | 170–182 |
| GCTTTCAATATATCATACGTGGT | ||||
| JcSSR7 | TTATCAACCTATCATCCTCAT | 53 | (TG)18 | 148–202 |
| GCACAGAATCAAACTCAAG | ||||
| JcSSR8 | ATTTAGCAGAACCCCAGAAC | 58 | (AC)17 | 154–182 |
| ATGTCTCTTTTCCATGTCCAA | ||||
| JcSSR9 | TCAACCCACCCTCATATAAACC | 60 | (AC)10 | 115 |
| TACACACTTGCCCTGATTTCTG | ||||
| JcSSR10 | TTCAGCCAGAATCTCAACAGT | 64 | (CT)16 | 178–184 |
| AGGAAGGATGGGAAGTGGGA | ||||
2.4. Genetic Diversity and Differentiation Statistics
3. Results
3.1. Genomic DNA Fingerprint Analysis By Means Of RAPD and Inter-SSR Markers
3.2. Genetic Diversity and Differentiation Statistics Based On SSR Markers
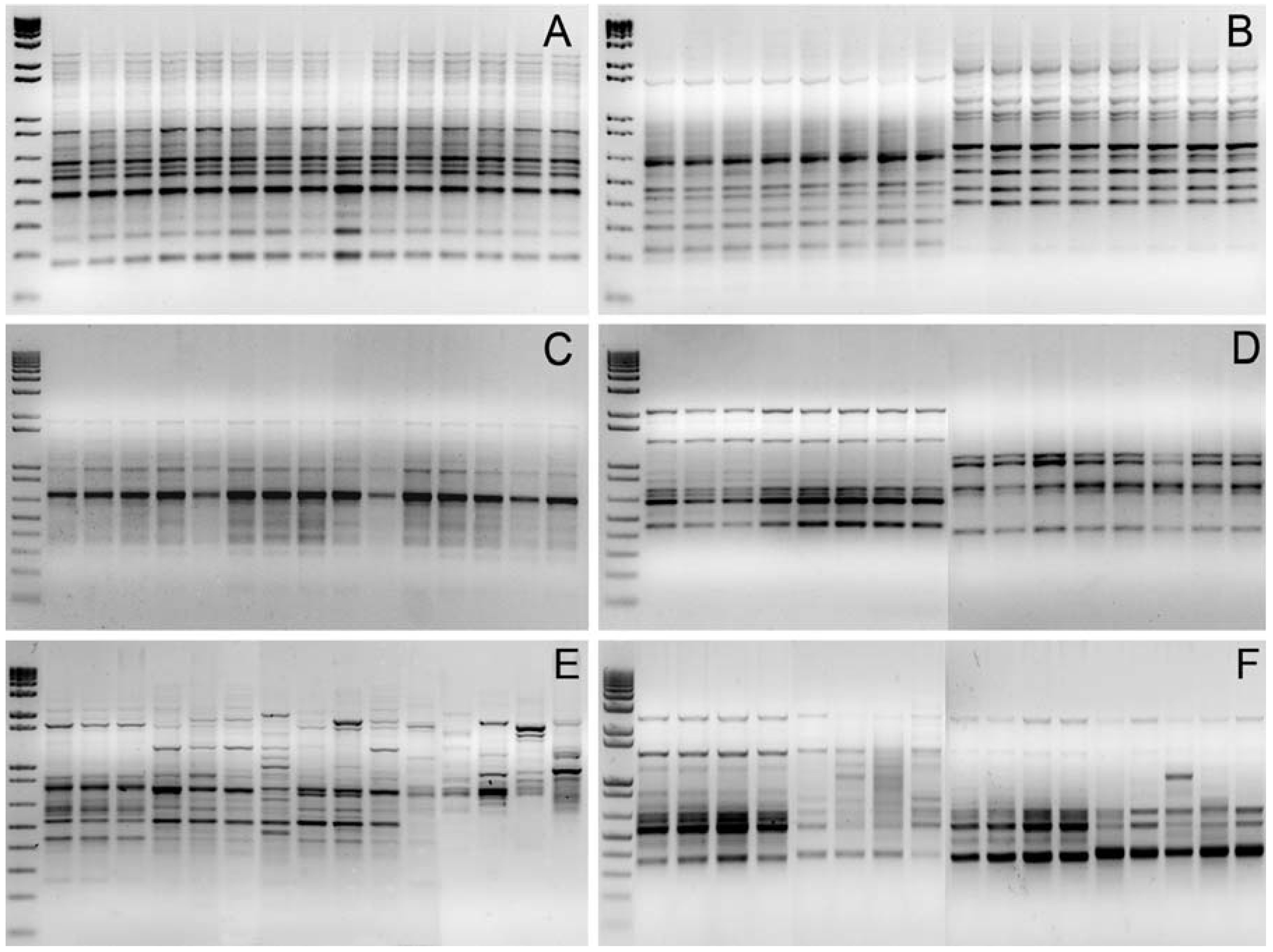
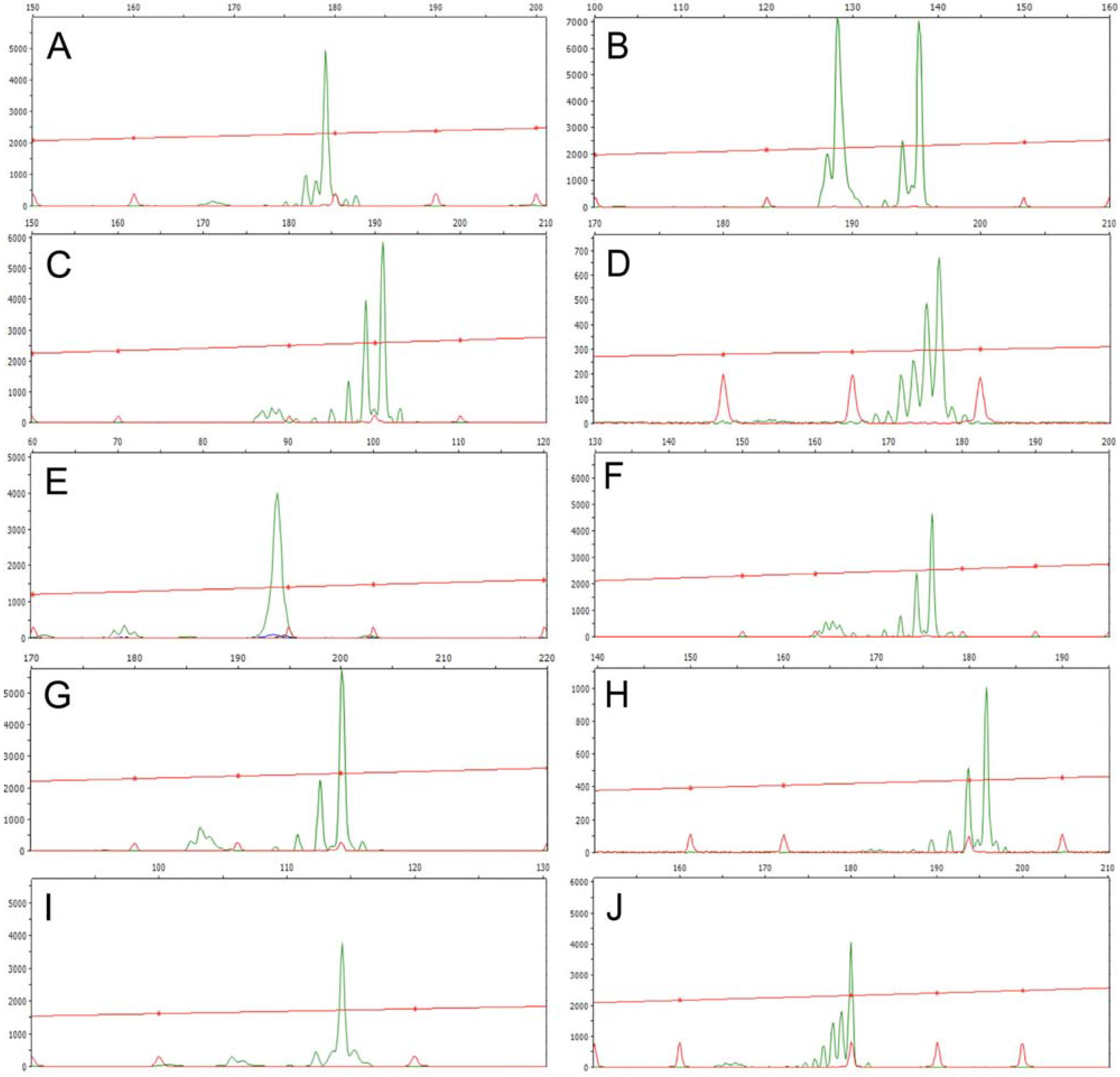
| Accession Id. | S | pi | no | ne | I | H | Fis |
|---|---|---|---|---|---|---|---|
| JcSSR1 | 106 | 0.8962 | 4 | 1.2372 | 0.4275 | 0.1917 | 0.7047 |
| JcSSR2 | 106 | 0.3962 | 4 | 3.2474 | 1.2656 | 0.6921 | −1.036 |
| JcSSR3 | 106 | 0.5943 | 6 | 2.5069 | 1.25 | 0.6011 | 0.4664 |
| JcSSR4 | 106 | 0.5472 | 6 | 2.7259 | 1.2407 | 0.6331 | 0.255 |
| JcSSR5 | 106 | 1 | 1 | 1 | 0 | 0 | 0 |
| JcSSR6 | 104 | 0.9231 | 4 | 1.1706 | 0.3512 | 0.1457 | 0.736 |
| JcSSR7 | 98 | 0.949 | 3 | 1.1082 | 0.227 | 0.0977 | 0.791 |
| JcSSR8 | 100 | 0.47 | 4 | 2.3397 | 0.9612 | 0.5726 | −0.5718 |
| JcSSR9 | 104 | 1 | 1 | 1 | 0 | 0 | 0 |
| JcSSR10 | 104 | 0.5962 | 5 | 2.0969 | 0.8902 | 0.5231 | −0.3234 |
| Mexico | 17 | 0.6611 | 2.9 | 2.0615 | 0.7485 | 0.4260 | 0.1138 |
| South America | 14 | 0.7857 | 2 | 1.5949 | 0.4407 | 0.2663 | −0.1609 |
| Africa | 30 | 0.8333 | 1.6 | 1.4241 | 0.3115 | 0.1978 | −0.4116 |
| Asia | 43 | 0.75 | 2 | 1.6789 | 0.481 | 0.2999 | 0.0998 |
| Mean | 104 | 0.7372 | 3.8 | 1.8433 | 0.6613 | 0.3457 | −0.1438 |
| St. Dev. | - | 0.2373 | 1.7512 | 0.8356 | 0.5168 | 0.2819 | - |
| Marker | Sample | H-statistics | F-statistics | Gene flow | ||||
|---|---|---|---|---|---|---|---|---|
| Locus Id. | Size | Ho | He | Hav | Fis | Fit | Fst | Nm |
| JcSSR1 | 106 | 0.0566 | 0.1936 | 0.1683 | 0.5048 | 0.6582 | 0.3096 | 0.56 |
| JcSSR2 | 106 | 0.717 | 0.6987 | 0.4832 | −0.4287 | 0.0339 | 0.3238 | 0.52 |
| JcSSR3 | 106 | 0.3208 | 0.6068 | 0.3985 | 0.1659 | 0.4413 | 0.3302 | 0.5 |
| JcSSR4 | 106 | 0.4717 | 0.6392 | 0.5684 | 0.067 | 0.2016 | 0.1443 | 1.48 |
| JcSSR5 | 106 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| JcSSR6 | 104 | 0.0385 | 0.1471 | 0.1665 | 0.7172 | 0.7415 | 0.0859 | 2.66 |
| JcSSR7 | 98 | 0.0204 | 0.0987 | 0.1354 | 0.6923 | 0.7808 | 0.2877 | 0.62 |
| JcSSR8 | 100 | 0.9 | 0.5784 | 0.5753 | −0.5078 | −0.4804 | 0.0182 | 13.52 |
| JcSSR9 | 104 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| JcSSR10 | 104 | 0.6923 | 0.5282 | 0.4794 | −0.5057 | −0.2708 | 0.156 | 1.35 |
| Mean | 104 | 0.3217 | 0.3491 | 0.2975 | −0.1438 | 0.0897 | 0.2042 | 0.97 |
| St. Dev. | - | 0.3494 | 0.2846 | 0.2274 | 0.4269 | 0.3567 | 0.1379 | 4.08 |
3.3. Ordination Analyses of Accessions and Population Structure Inferences
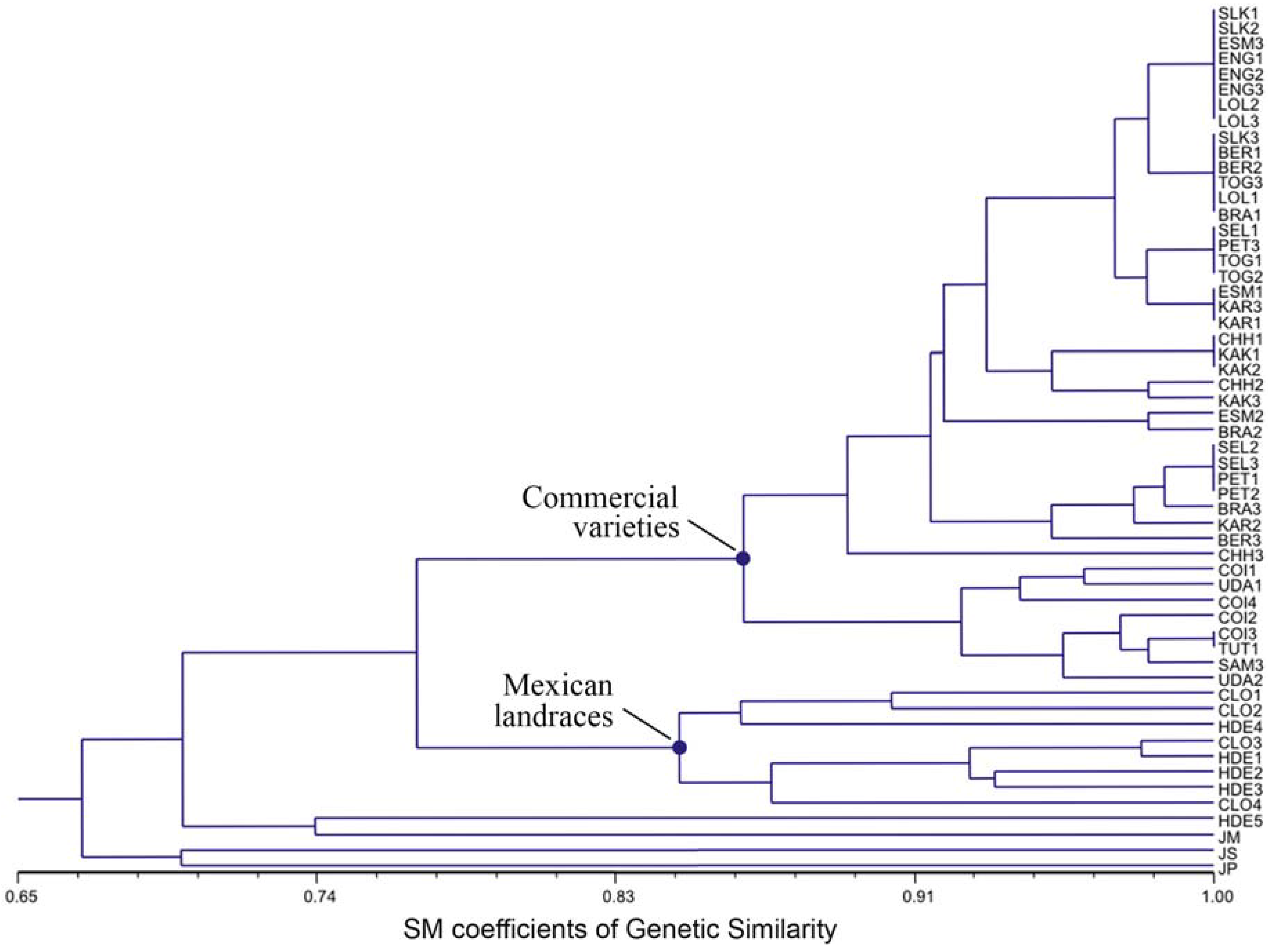
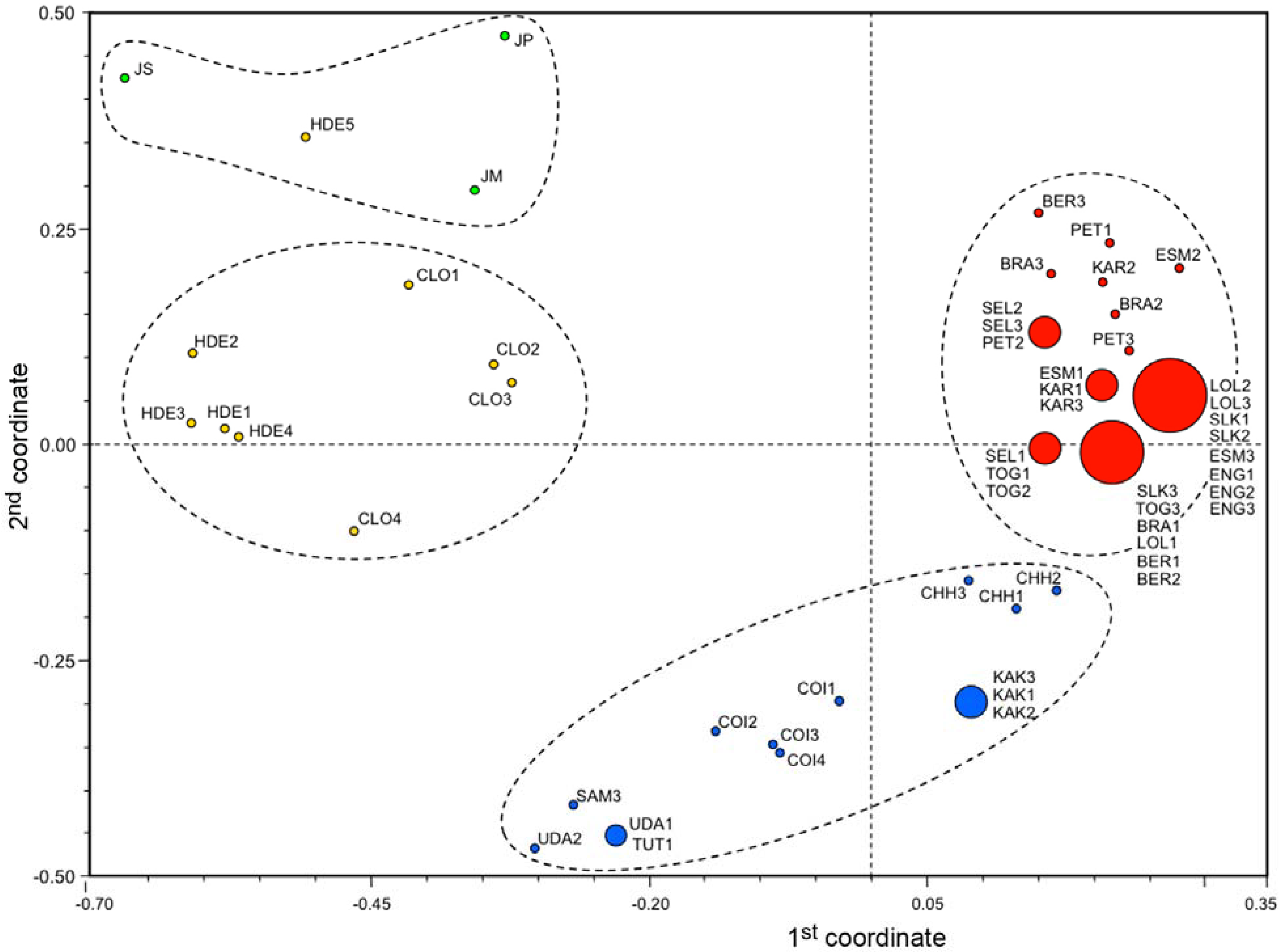

| Geographical | Inferred clusters | No. | |||
|---|---|---|---|---|---|
| origin | 1: varieties | 2: varieties | 3: landraces | 4: outgroups | accessions |
| Mexico (1) | 0.012 | 0.007 | 0.642 | 0.339 | 12 |
| South America (2) | 0.844 | 0.149 | 0.004 | 0.004 | 7 |
| Africa (3) | 0.982 | 0.011 | 0.004 | 0.003 | 15 |
| Asia (4) | 0.983 | 0.01 | 0.004 | 0.004 | 9 |
| India (5) | 0.06 | 0.924 | 0.004 | 0.012 | 13 |
3.4. FCSS Analysis for the Estimation of Seed DNA Contents
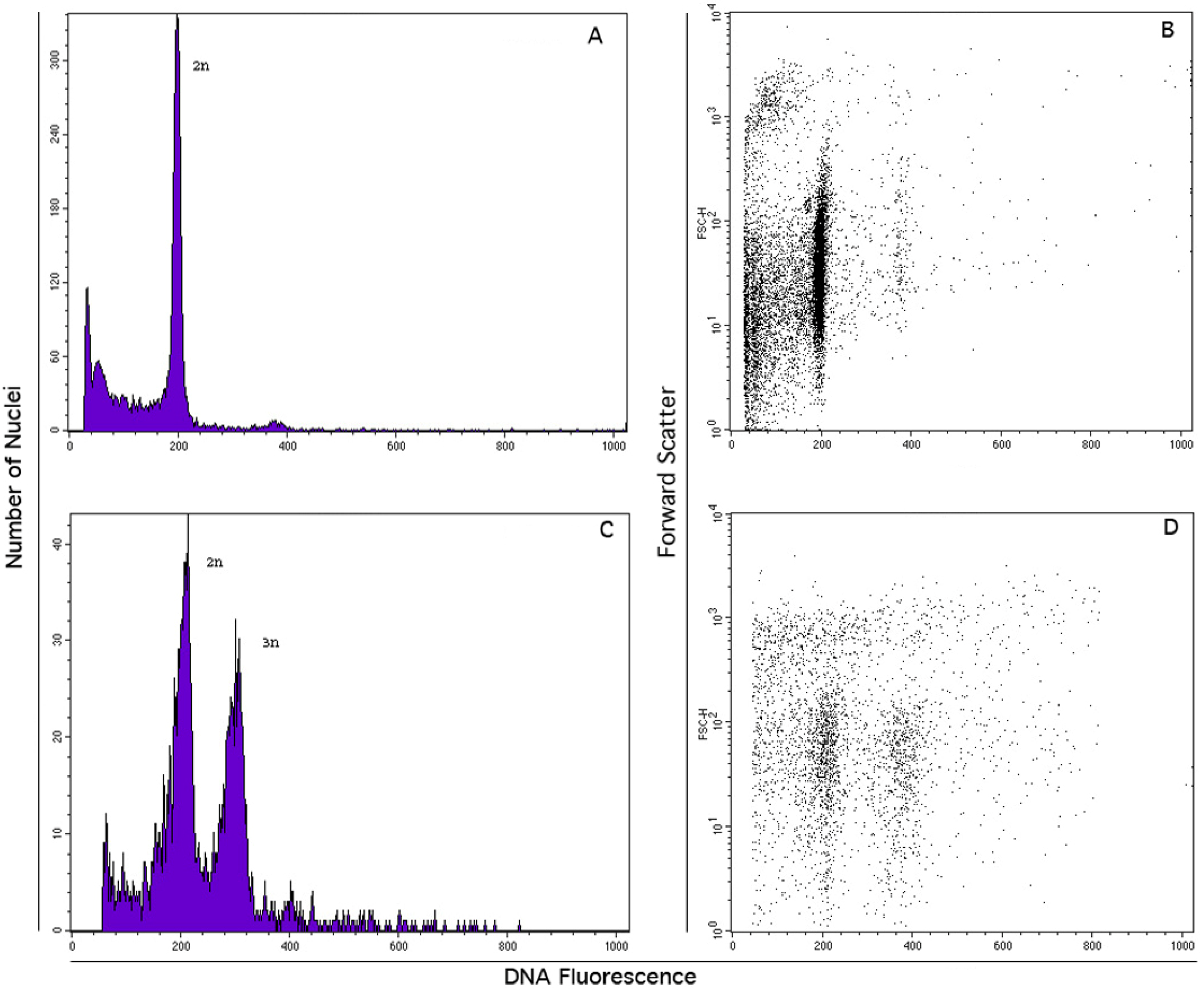

| Accession Id. | No. seeds | 2x embryo | 3x endosperm |
|---|---|---|---|
| JcCLO | 4 | 4 | 4 |
| JcHDE | 5 | 5 | 5 |
| JcBRA | 2 | 1 | 1 |
| JcSAM | 1 | 1 | 1 |
| JcLOL | 25 | 21 | 12 |
| JcTOG | 2 | 2 | 2 |
| JcENG | 20 | 19 | 18 |
| JcBER | 22 | 19 | 8 |
| JcESM | 23 | 12 | 5 |
| JcKAR | 2 | 1 | 1 |
| JcSLK | 22 | 21 | 3 |
| JcPET | 22 | 17 | 5 |
| JcSEL | 22 | 19 | 10 |
| JcJOR | 2 | 1 | 1 |
| JcCHH | 2 | 1 | 1 |
| JcUDP | 2 | 1 | 1 |
| JcKAK | 5 | 4 | 3 |
| JcTUT | 4 | 4 | 4 |
| JcCOI | 3 | 2 | 2 |
| 190 | 155 | 87 |
4. Discussion
4.1. Analysis of Plant DNA Markers and Seed DNA Contents Shed Light on the Genetic Diversity and Reproductive Biology of J. curcas
5. Conclusions
References
- Zieler, H.; Richardson, T.; Schwartz, A.; Herrgard, M.; Lomelin, D.; Mathur, E.; Cheah, S.C.; Tee, T.S.; Lee, W.W.; Chua, K.L.; Kwan, B.H.; Miller, J.; Koren, S.; Chan, A.P.; Maiti, R.; Amedeo, P.; Ferriera, S.; Rogers, Y.H.; Sutton, G.; Town, C.D.; Venter, J.C. Whole-genome shotgun sequencing of the oil palm and Jatropha genomes. In Proceedings of the Plant and Animal Genomes XVIII Conference; Sherago International, Inc.: San Diego, CA, USA, January 2010; p. W. 360. [Google Scholar]
- Nature Editorial. Kill king corn. Nature 2007, 449, 637. [Google Scholar]
- Foidl, N.; Foidl, G.; Sanchez, M.; Mitelbach, M.; Heckel, S. Jatropha curcas L. as a source for the production of biofuel in Nicaragua. Bioresource Technol. 1996, 58, 77–82. [Google Scholar] [CrossRef]
- Gubitz, G.M.; Mittelbach, M.; Trabi, M. Exploitation of the tropical oil seed plant Jatropha curcas L. Bioresource Technol. 1999, 67, 73–82. [Google Scholar] [CrossRef]
- Francis, G.; Edinger, R.; Becker, K. A concept for simultaneous wasteland reclamation, fuel production, and socio- econimic development in degraded areas in India: Need potential and prospectives of Jatropha plantations. Nat. Resour. Forum. 2005, 29, 12–24. [Google Scholar] [CrossRef]
- Berchmans, H.J.; Hirata, S. Biodisel production from crude Jatropha curcas L. seed oil with a high content of free fatty acids. Bioresource Technol. 2008, 99, 1716–1721. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, S. An evaluation of multipropose oil seed crop for industrial uses (Jatropha curcas): A review. Ind. Crops Prod. 2008, 28, 1–10. [Google Scholar] [CrossRef]
- King, R.D.; Rowland, J.; Oliver, S.G.; Young, M.; Aubrey, W.; Byrne, E.; Liakata, M.; Markham, M.; Pir, P.; Soldatova, L.N.; Sparkes, A.; Whelan, K.E.; Clare, A. The automation of science. Science 2009, 324, 85–89. [Google Scholar] [CrossRef]
- Basha, S.D.; Francis, G.; Makkar, H.P.S.; Becker, K.; Sujatha, M. A comparative study of biochemical traits and molecular markers for assessment of genetic relationships between Jatropha curcas L. germplasm from different countries. Plant Sci. 2009, 176, 812–823. [Google Scholar] [CrossRef]
- Ambrosi, D.G.; Galla, G; Purelli, M.; Barbi, T; Fabbri, A.; Barcaccia, G. DNA markers and FCSS analyses shed light on the genetic diversity and reproductive biology of Jatropha curcas L. In Proceedings of the Plant and Animal Genomes XVIII Conference; Sherago International, Inc.: San Diego, CA, USA, January 2010; p. 129. [Google Scholar]
- Spillane, C.; Curtis, M.D.; Grossniklaus, U. Apomixis technology development. Virgin births in farmers fields. Nat. Biotechnol. 2004, 22, 687–691. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Datta, K.; Datta, S.K. Floral biology, floral resource constraints and pollination limitation in Jatropha curcas L. Pakistan J. Biological Sci. 2005, 8, 456–460. [Google Scholar] [CrossRef]
- Luo, C-W.; Li, K.; Chen, Y.; Sun, Y.Y. Floral display and breeding system of Jatropha curcas L. Forestry Stud. China 2007, 9, 114–119. [Google Scholar] [CrossRef]
- Popluechai, S.; Breviario, D.; Mulpuri, S.; Makkar, H.P.S.; Raorane, M.; Reddy, A.R.; Palchetti, E.; Gatehouse, A.M.R.; Syers, J.K.; O’Donnell, A.G.; Kohli, A. Narrow genetic and apparent phenetic diversity in Jatropha curcas: Initial success with generating low phorbol ester interspecific hybrids. Nature Proceedings 2009. (available at hdl: 10101/npre.2009.2782.1). [Google Scholar]
- Pamidimarri, D.V.N.S.; Singh, S.; Mastan, S.G.; Patel, J.; Reddy, M.P. Molecular characterization and identification of markers for toxic and non-toxic varieties of Jatropha curcas L. using RAPD, AFLP, and SSR markers. Mol. Biol. Rep. 2009, 36, 1357–1364. [Google Scholar] [CrossRef]
- Basha, S.D.; Sujatha, M. Inter and intra-population variability of Jatropha curcas L. characterized by RAPD and ISSR markers and development of population-specific SCAR markers. Euphytica 2007, 156, 375–386. [Google Scholar] [CrossRef]
- Yu, C.; Wu, G.; Peng, J. ISSR-based genetic diversity of Jatropha curcas germplasm in China. In Proceedings of the Plant and Animal Genomes XVIII Conference; Sherago International, Inc.: San Diego, CA, USA, January 2010; p. 212. [Google Scholar]
- Pamidimarri, D.V.N.S.; Nirali, P.; Reddy, M.P.; Radhakrishnan, T. Comparative study of interspecific divergence and phylogenic analysis of genus Jatropha by RAPD and AFLP. Mol. Bio. Rep. 2008, 36, 901–907. [Google Scholar]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef]
- Kaushik, N.; Kumar, K.; Kumar, S.; Kaushik, N.; Roy, S. Genetic variability and divergence studies in seed traits and oil content of Jatropha (Jatropha curcas L.) accessions. Biomass and Bioenergy 2007, 31, 497–502. [Google Scholar] [CrossRef]
- Popluechai, S.; Raorane, M.; Syers, K.J.; O’Donnell, A.G.; Kohli, A. Research needs to make Jatropha a viable alternate oilseed crop. In Proceedings of the International workshop on Feasibility of non-edible oil seed crops for biofuel production, Mae Fah Luang University; Syers, K.J., Wood, D., Eds.; MFLU publication: Chiang Rai, Thailand, May 2007; pp. 23–38. [Google Scholar]
- Gressel, J. Transgenics are imperative for biofuel crops. Plant Sci. 2008, 174, 246–263. [Google Scholar] [CrossRef]
- Matzk, F.; Meister, A.; Schubert, I. An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J. 2000, 21, 97–108. [Google Scholar] [CrossRef]
- Dolezel, J.; Binarova, P.; Lucretti, S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant. 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Mendonça, M.A.C.; Carvalho, C.R.; Clarindo, R.W. DNA content differences between male and female chicken (Gallus gallus domesticus) nuclei and Z and W chromosomes resolved by image. J. Histochem. Cytochem. 2009, 58, 229–235. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; NY Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; p. 1659. [Google Scholar]
- Barcaccia, G.; Mazzucato, A.; Belardinelli, A.; Pezzotti, M.; Lucretti, S.; Falcinelli, M. Inheritance of parental genomes in progenies of Poa pratensis L. from sexual and apomictic genotypes as assessed by RAPD markers and flow cytometry. Theor. Appl. Genet. 1997, 95, 516–524. [Google Scholar] [CrossRef]
- Barcaccia, G.; Lucchin, M.; Parrini, P. Characterization of a flint maize (Zea mays var. indurata) Italian landrace: II. Genetic diversity and relatedness assessed by PCR-based molecular markers. Genet. Res. Crop Evol. 2003, 50, 253–271. [Google Scholar] [CrossRef]
- Barcaccia, G.; Meneghetti, S.; Albertini, E.; Triest, L.; Lucchin, M. Linkage mapping in tetraploid willows: segregation of molecular markers and estimation of linkage phases support an allotetraploid structure for Salix alba x Salix fragilis interspecific hybrids. Heredity 2003, 90, 169–180. [Google Scholar] [CrossRef]
- Hayden, M.J.; Nguyen, T.M.; Watrman, A.; McMichael, G.L.; Chalmers, K.J. Application of multiplex-ready PCR for fluorescence-based SSR genotyping in barley and wheat. Mol. Breeding 2008, 21, 271–281. [Google Scholar] [CrossRef]
- Zubieta, C.G.; Ghiselli, L.; Benedettelli, S.; Palchetti, E. Development of novel SSR markers from genomic microsatellite library in Jatropha curcas L. In Proceedings of the 53rd Italian Society of Agricultural Genetics Annual Congress; SIGA publisher: Torino, Italy, September 2009; pp. 7–60. [Google Scholar]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Lewontin, R.C. The Genetic Basis of Evolutionary Change; Columbia University Press: New York, NY, USA, 1974. [Google Scholar]
- McDermott, J.M.; McDonald, B.A. Gene flow in plant pathosystems. Annu. Rev. Phytopathol. 1993, 31, 353–373. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F-statistics with special regards to systems of matings. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations. Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978; vol. 4. [Google Scholar]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar]
- Goodman, S.J. Rst Calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and determining their significance. Mol. Ecol. 1997, 6, 881–885. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE version 1.21; CIFOR and University of Alberta: Edmonton, Canada, 1997. [Google Scholar]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy; Freeman W.H. Co: San Francisco, CA, USA, 1973. [Google Scholar]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1996, 53, 325–338. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy System, (version 2.1); Exeter Publishing, Ltd.: Setauket, NY, USA, 2002. [Google Scholar]
- Pritchard, J.K.; Stephens, P.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Carvalho, C.R.; Clarindo, W.R.; Prac, M.M.; Araujo, F.S.; Carels, N. Genome size, base composition and karyotype of Jatropha curcas L., an important biofuel plant. Plant Sci. 2008, 174, 613–617. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar]
- Loureiro, J.; Rodriguez, E.; Dolezel, J.; Santos, C. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Annals Botany 2006, 98, 679–689. [Google Scholar] [CrossRef]
- Savidan, Y.H. Apomixis: Genetics and breeding. Plant Breed. 2000, 18, 10–86. [Google Scholar]
- Barcaccia, G.; Baumlein, H.; Sharbel, T.F. Apomixis in St. John’s wort: An overview and glimpse towards the future. In Apomixis: Evolution, Mechanisms and Perspectives; Hörandle, E., Grossniklaus, U., Van Dijk, P., Sharbel, T.F., Eds.; Intl. Association of Plant Taxonomy - Koeltz Scientific Books: Vienna, Austria, 2007; Chapter XIV; pp. 259–280. [Google Scholar]
- Asker, S.E.; Jerling, L. Apomixis in plants; CRC Press: London, UK, 1992. [Google Scholar]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer-Verlag: Berlin, Germany, 1984; pp. 475–518. [Google Scholar]
- Allard, R.W. History of plant population genetics. Annu. Rev. Genet. 1999, 33, 1–27. [Google Scholar] [CrossRef]
- Heller, J. Physic Nut. Jatropha curcas L. Promoting the Conservation and Use of Underutilized and Neglected Crops; Institute of Plant Genetics and Crop Plant Research (IPK), International Plant Genetic Resources Institute (IPGRI): Gatersleben, Germany, Rome, Italy, 1996; p. 65. [Google Scholar]
- Jongshaap, R.E.E.; Corré, W.J.; Bindraban, P.S.; Brandenburg, W.A. Claims and Facts on Jatropha curcas L. Global Jatropha curcas Evaluation, Breeding and Propagation Programme. Report 158; Plant Research International B.V., Wageningen Stichting Het Groene Woudt: Laren, The Netherlands, 2007. [Google Scholar]
- Montes Osorio, L.R.; Van Loo, E.N.R.; Jongshaap, R.E.E.; Visser, R.G.H.; Azurdia, C. Global Jatropha curcas Genetic Diversity Study and its Application in Breeding Programs. In JatrophaWorld 2008: An OUTSTANDING Event – Two Thumbs Up, Miami, FL, USA, June 2008.
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ambrosi, D.G.; Galla, G.; Purelli, M.; Barbi, T.; Fabbri, A.; Lucretti, S.; Sharbel, T.F.; Barcaccia, G. DNA Markers and FCSS Analyses Shed Light on the Genetic Diversity and Reproductive Strategy of Jatropha curcas L. Diversity 2010, 2, 810-836. https://doi.org/10.3390/d2050810
Ambrosi DG, Galla G, Purelli M, Barbi T, Fabbri A, Lucretti S, Sharbel TF, Barcaccia G. DNA Markers and FCSS Analyses Shed Light on the Genetic Diversity and Reproductive Strategy of Jatropha curcas L. Diversity. 2010; 2(5):810-836. https://doi.org/10.3390/d2050810
Chicago/Turabian StyleAmbrosi, Daria Gigliola, Giulio Galla, Marina Purelli, Tommaso Barbi, Andrea Fabbri, Sergio Lucretti, Timothy Francis Sharbel, and Gianni Barcaccia. 2010. "DNA Markers and FCSS Analyses Shed Light on the Genetic Diversity and Reproductive Strategy of Jatropha curcas L." Diversity 2, no. 5: 810-836. https://doi.org/10.3390/d2050810






