The Polyextreme Ecosystem, Salar de Huasco at the Chilean Altiplano of the Atacama Desert Houses Diverse Streptomyces spp. with Promising Pharmaceutical Potentials
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Actinomyces
2.2. Identification of Bacterial Isolates
2.2.1. Morphological Characterization Based on Color Grouping
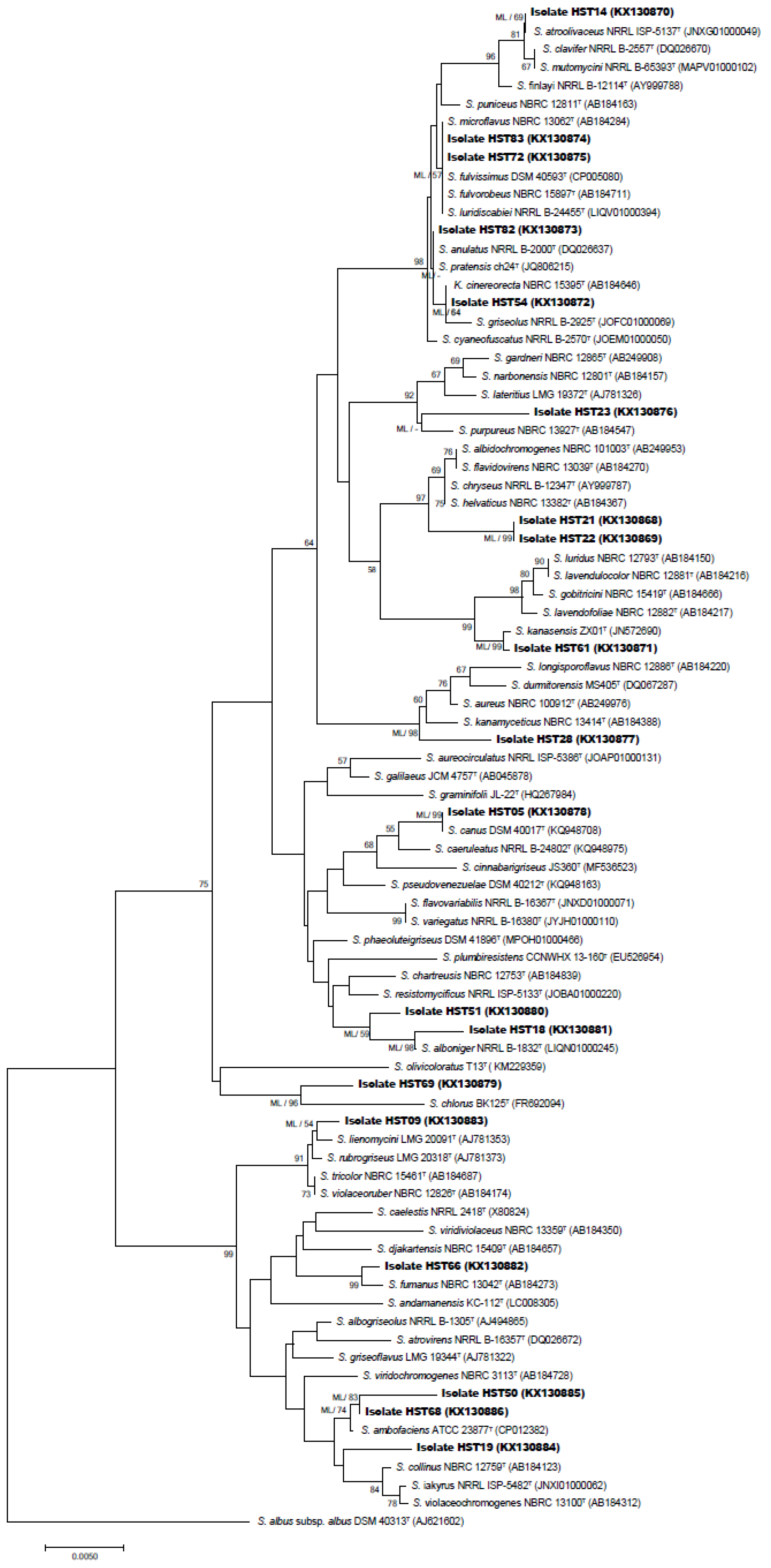
2.2.2. 16S rRNA Gene-Based Phylogenetic Analysis
2.3. Evaluations of Pharmaceutical Potentials
2.3.1. Genotyping of Nonribosomal Peptide Synthetase (NRPS)
2.3.2. Preparation of Crude Extracts and Bioactivity Assessments
2.3.3. Profiling of Metabolites in Crude Extracts
3. Results and Discussion
3.1. Selective Isolation and Identification of the Isolates
3.2. Secondary Metabolite Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Watve, M.; Tickoo, R.; Jog, M.; Bhole, B. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Anargyros Chaliotis, A.; Mossialos, D. Discovery Strategies of Bioactive Compounds Synthesized by Nonribosomal Peptide Synthetases and Type-I Polyketide Synthases Derived from Marine Microbiomes. Mar. Drugs 2016, 14, 80. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Goodfellow, M. Biosystematics of alkaliphilic Streptomycetes isolated from seven locations across a beach and dune sand system. Antonie Leeuwenhoek 2008, 94, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Stach, J.E.; Ward, A.C.; Goodfellow, M. Marine actinobacteria: Perspectives, challenges, future directions. Antonie Leeuwenhoek 2005, 87, 65–79. [Google Scholar] [CrossRef]
- Pathom-aree, W.; Stach, J.E.M.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Diversity of Actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 2006, 10, 181–189. [Google Scholar] [CrossRef]
- Lyutskanova, D.; Ivanova, V.; Stoilova-Disheva, M.; Kolarova, M.; Aleksieva, K.; Peltekova, V. Isolation and characterization of a psychrotolerant Streptomyces strain from permafrost soil in Spitsbergen, producing phtalic acid ester. Biotechnol. Biotechnol. Equip. 2009, 23, 1220–1224. [Google Scholar] [CrossRef]
- Steven, B.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 2008, 10, 3388–3403. [Google Scholar] [CrossRef] [Green Version]
- Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Ferguson, G.; Bull, A.T.; Goodfellow, M.; Ebel, R.; Jaspars, M. Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J. Nat. Prod. 2011, 74, 1491–1499. [Google Scholar] [CrossRef]
- Fiedler, H.P.; Bruntner, C.; Bull, A.T.; Ward, A.C.; Goodfellow, M.; Potterat, O.; Puder, C.; Mihm, G. Marine Actinomycetes as a source of novel secondary metabolites. Antonie Leeuwenhoek 2005, 87, 37–42. [Google Scholar] [CrossRef]
- Goodfellow, M.; Nouioui, I.; Sanderson, R.; Xie, F.; Bull, A.T. Rare taxa and dark microbial matter: Novel bioactive actinobacteria abound in Atacama Desert soils. Antonie Leeuwenhoek 2018, 111, 1315–1332. [Google Scholar] [CrossRef]
- Santhanam, R.; Okoro, C.K.; Rong, X.; Huang, Y.; Bull, A.T.; Weon, H.Y.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M. Streptomyces atacamensis sp. nov., isolated from an extreme hyper-arid soil of the Atacama Desert, Chile. Int. J. Syst. Evol. Microbiol. 2012, 62, 2680–2684. [Google Scholar] [CrossRef]
- Santhanam, R.; Okoro, C.K.; Rong, X.; Huang, Y.; Bull, A.T.; Andrews, B.A.; Asenjo, J.A.; Weon, H.Y.; Goodfellow, M. Streptomyces deserti sp. nov., isolated from hyper-arid Atacama Desert soil. Antonie Leeuwenhoek 2012, 101, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, R.; Rong, X.; Huang, Y.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M. Streptomyces bullii sp. nov., isolated from a hyper-arid Atacama Desert soil. Antonie Leeuwenhoek 2013, 103, 367–373. [Google Scholar] [CrossRef]
- Busarakam, K.; Bull, A.T.; Girard, G.; Labeda, D.P.; Van Wezel, G.P.; Goodfellow, M. Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie Leeuwenhoek 2014, 105, 849–861. [Google Scholar] [CrossRef]
- Rateb, M.E.; Houssen, W.E.; Harrison, W.T.A.; Deng, H.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; et al. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J. Nat. Prod. 2011, 74, 1965–1971. [Google Scholar] [CrossRef]
- Nachtigall, J.; Kulik, A.; Helaly, S.; Bull, A.T.; Goodfellow, M.; Asenjo, J.A.; Maier, A.; Wiese, J.; Imhoff, J.F.; Sussmuth, R.D.; et al. Atacamycins A–C, 22-membered antitumor macrolactones produced by Streptomyces sp. C38. J. Antibiot. 2011, 64, 775–780. [Google Scholar] [CrossRef]
- Elsayed, S.S.; Trusch, F.; Deng, H.; Raab, A.; Prokes, I.; Busarakam, K.; Asenjo, J.A.; Andrews, B.A.; Van West, P.; Bull, A.T.; et al. Chaxapeptin, a lasso peptide from extremotolerant Streptomyces leeuwenhoekii strain C58 from the hyperarid Atacama Desert. J. Org. Chem. 2015, 80, 10252–10260. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Beese, P.; Ohlendorf, B.; Erhard, A.; Zinecker, H.; Dorador, C.; Imhoff, J.F. Abenquines A–D: Aminoquinone derivatives produced by Streptomyces sp. strain DB634. J. Antibiot. 2011, 64, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Dorador, C.; Vila, I.; Witzel, K.-P.; Imhoff, J.F. Bacterial and archaeal diversity in high altitude wetlands of the Chilean Altiplano. Fundam. Appl. Limnol. 2013, 182, 135–159. [Google Scholar] [CrossRef]
- Molina, V.; Hernandez, K.; Dorador, C.; Hengst, M.; Pérez, V. Bacterial active community cycling in response to solar radiation and their influence on nutrient changes in a high-altitude wetland. Front. Microbiol. 2016, 7, 1823. [Google Scholar] [CrossRef]
- Dorador, C.; Vila, I.; Remonsellez, F.; Imhoff, J.F.; Witzel, K.-P. Unique clusters of Archaea in Salar de Huasco, an athalassohaline evaporitic basin of the Chilean Altiplano. FEMS Microbiol. Ecol. 2010, 73, 291–302. [Google Scholar] [CrossRef]
- Dorador, C.; Meneses, D.; Urtuvia, V.; Demergasso, C.; Vila, I.; Witzel, K.P.; Imhoff, J.F. Diversity of Bacteroidetes in high-altitude saline evaporitic basins in northern Chile. J. Geophys. Res. 2009, 114, G00D05. [Google Scholar] [CrossRef]
- Kuester, E.; Williams, S.T. Selection of media for isolation of Streptomycetes. Nature 1964, 202, 928–929. [Google Scholar] [CrossRef]
- Vickers, J.C.; Williams, S.T.; Ross, G.W. A taxonomic approach to selective isolation of Streptomycetes from soil. In Biological and Biochemical Aspects of Actinomycetes; Ortiz-Ortiz, L., Bojalil, L.F., Yakoleff, V., Eds.; Academic Press: Orlando, FL, USA, 1984; pp. 553–561. [Google Scholar]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Kelly, K.L. Color Name Charts Illustrated with Centroid Colors; Supplement to NBS Circular 553. Inter-Society Color Council—National Bureau of Standards (ISCC-NBS); U. S. National Bureau of Standards: Washington, DC, USA, 1964.
- Stackebrandt, E.; Liesack, W. Nucleic Acids and Classification. In Handbook of New Bacterial Systematics; Goodfellow, M., O’Donnell, A.G., Eds.; Academic Press: London, UK, 1993; pp. 151–194. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M. Cole JR (2007) Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 1984, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Ayuso-Sacido, A.; Genilloud, O. New PCR primers for the screening of NRPS and PKS-I systems in Actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a Cytotoxic Polyketide from a Streptomyces Strain Isolated from the Marine Sponge Halichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Ohlendorf, B.; Labes, A.; Erhard, A.; Imhoff, J.F. Calcarides A–E, antibacterial macrocyclic and linear polyesters from a calcarisporium strain. Mar. Drugs 2013, 11, 3309–3323. [Google Scholar] [CrossRef]
- Buckingham, J. Dictionary of Natural Products on DVD, version 21:1; CRC Press Taylor and Francis Group: London, UK, 2012. [Google Scholar]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [Green Version]
- Lindenbein, W. Über Einige Chemisch Interessante Aktinomycetenstämme Und Ihre Klassifizierung. Arch. Mikrobiol. 1952, 17, 361–383. [Google Scholar] [CrossRef]
- De Querioz, V.M.; Albert, C.A. Streptomyces iakyrus nov. sp., produtor dos antibióticos Iaquirina I, IIe, III. Rev. Inst. Antibiot. Univ. Recife 1962, 4, 33–46. [Google Scholar]
- Ryabova, I.D.; Preobrazhenskaya, T.P. Problems of Classification of Actinomycetes-Antagonists; Gauze, G.F., Preobrazhenskaya, T.P., Kudrina, E.S., Blinov, N.O., Ryabova, I.D., Sveshnikova, M.A., Eds.; Government Publishing House of Medical Literature: Medgiz, Moscow, Russia, 1957; pp. 1–398.
- Gause, G.F.; Preobrazhenskaya, T.P.; Sveshnikova, M.A.; Terekhova, L.P.; Maximova, T.S. A Guide for the Determination of Actinomycetes. Genera Streptomyces, Streptoverticillium, and Chainia; Nauka: Moscow, Russia, 1983. [Google Scholar]
- Han, L.; Zhang, G.; Miao, G.; Zhang, X.; Feng, J. Streptomyces kanasensis sp. nov., an Antiviral Glycoprotein Producing Actinomycete Isolated from Forest Soil Around Kanas Lake of China. Curr. Microbiol. 2015, 71, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Okami, Y.; Umezawa, H. Production and isolation of a new antibiotic, kanamycin. J. Antibiot. 1957, 10, 181–189. [Google Scholar]
- Goodfellow, M.; Williams, S.T.; Alderson, G. Transfer of Kitasatoa purpurea Matsuma and Hata to the genus Streptomyces as Streptomyces purpurea comb. nov. Syst. Appl. Microbiol. 1986, 8, 65–66. [Google Scholar] [CrossRef]
- Onaka, H. Biosynthesis of heterocyclic antibiotics in Actinomycetes and an aproach to synthesize the natural compounds. Actinomycetologica 2006, 20, 62–71. [Google Scholar] [CrossRef]
- Sánchez, C.; Méndez, C.; Salas, J.A. Engineering biosynthetic pathways to generate antitumor indolocarbazole derivatives. J. Ind. Microbiol. Biotechnol. 2006, 33, 560–568. [Google Scholar] [CrossRef]
- Bowers, K.J.; Mesbah, N.M.; Wiegel, J. Biodiversity of poly-extremophilic Bacteria: Does combining the extremes of high salt, alkaline pH and elevated temperature approach a physico-chemical boundary for life? Saline Syst. 2009, 5, 9. [Google Scholar] [CrossRef]
- Dorador, C.; Vila, I.; Imhoff, J.F.; Witzel, K.-P. Cyanobacterial diversity in Salar de Huasco, a high altitude saline wetland in northern Chile: An example of geographical dispersion? FEMS Microbiol. Ecol. 2008, 64, 419–432. [Google Scholar] [CrossRef] [PubMed]

| Class | Family | Genus | Number of Isolate (%) | Site of Isolation |
|---|---|---|---|---|
| Actinobacteria | Streptomycetaceae | Streptomyces | 65 (86%) | H0 and H6 |
| Nocardiopsaceae | Nocardiopsis | 7 (9%) | H0 | |
| Micromonosporaceae | Micromonospora | 2 (3%) | H0 | |
| Bacilli | Bacillaceae | Bacillus | 1 (1%) | H0 |
| Gammaproteobacteria | Pseudomonadaceae | Pseudomonas | 1 (1%) | H0 |
| Isolate Code | Site of Isolation | Morphological Characteristics b | Molecular Characteristics | |||||
|---|---|---|---|---|---|---|---|---|
| Aerial Spore Mass | Substrate Mycelium | Diffusible Pigments | Closest Type Strain | 16 rRNA Gene Accession Number | Similarity (%) | NRPS (Adenylation Domain) | ||
| HST05 | H6 | White | Light yellowish pink | - | Streptomyces canus DSM 40017T | KQ948708 | 96.8 | + |
| HST09 | H6 | Dark greyish blue | Deep purplish red | - | Streptomyces lienomycini LMG 20091T | AJ781353 | 99.6 | + |
| HST14 | H6 | White | Pale violet | Light greyish brown | Streptomyces atroolivaceus NRRL ISP-5137T | JNXG01000049 | 96.0 | - |
| HST18 | H6 | - | Dark olive green | Moderate yellow green | Streptomyces alboniger NRRL B-1832T | LIQN01000245 | 96.7 | + |
| HST19 | H6 | Dark greyish blue | Dark greenish yellowish green | Dark greyish yellowish brown | Streptomyces collinus NBRC 12759T | AB184123 | 99.2 | + |
| HST21 | H6 | White | Greyish yellow | Moderate olive brown | Streptomyces albidochromogenes NBRC 101003T | AB249953 | 99.1 | + |
| HST22 | H6 | White | Deep orange yellow | Moderate olive brown | Streptomyces albidochromogenes NBRC 101003T | AB249953 | 99.0 | + |
| HST23 | H6 | Light pink | Light orange yellow | Dark yellow | Streptomyces purpureus NBRC 13927T | AB184547 | 98.8 | - |
| HST28 | H6 | White | Light olive brown | Dark brown | Streptomyces kanamyceticus NBRC 13414T | AB184388 | 98.8 | + |
| HST50 | H6 | White | Greyish reddish brown | - | Streptomyces ambofaciens ATCC 23877T | CP012382 | 96.0 | - |
| HST51 | H6 | Light pink | Strong yellowish brown | - | Streptomyces alboniger NRRL B-1832T | LIQN01000245 | 98.6 | + |
| HST54 | H6 | White | Deep red | Light greyish brown | Streptomyces griseolus NRRL B-2925T | JOFC01000069 | 97.6 | - |
| HST61 | H6 | Pale yellowish pink | Pale orange yellow | - | Streptomyces kanasensis ZX01T | JN572690 | 99.6 | - |
| HST66 | H6 | White | Deep greenish yellow | - | Streptomyces fumanus NBRC 13042T | AB184273 | 96.9 | + |
| HST68 | H6 | Greyish blue | Pinkish grey | - | Streptomyces ambofaciens ATCC 23877T | CP012382 | 96.0 | + |
| HST69 | H0 | White | Brilliant yellow | - | Streptomyces chlorus BK125T | LIQN01000245 | 98.1 | + |
| HST72 | H0 | White | Strong yellowish brown | Strong brown | Streptomyces microflavus NBRC 13062T | AB184284 | 97.9 | - |
| HST82 | H0 | White | Pale yellowish pink | - | Streptomyces cyaneofuscatus NRRL B-2570T | JOEM01000050 | 95.6 | - |
| HST83 | H0 | White | Strong yellowish brown | Strong brown | Streptomyces pratensis ch24T | JQ806215 | 97.8 | + |
| Crude Extract Derived From Streptomyces Isolate Grown in Different Media | Growth Inhibition (%) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibacterial Activity | Antifungal Activity | Cytotoxicity | |||||||||
| Gram-Positive Bacteria | Gram-Negative Bacteria | ||||||||||
| Se | MRSA | Pa | Xc | Ea | Ca | Tru | Sep | PhA | NIH-3T3 | HepG2 | |
| HST09-GYM | 96 | 92 | - | 27 | - | - | 72 | - | - | 99 | 79 |
| HST09-SPM | 28 | 28 | - | - | - | - | - | - | - | 36 | - |
| HST14-GYM | - | - | - | - | - | - | 33 | - | - | 50 | 40 |
| HST14-SPM | - | - | - | - | - | - | 30 | - | - | 51 | 42 |
| HST19-GYM | - | - | 57 | 55 | - | - | 58 | - | - | 57 | 34 |
| HST19-SPM | 61 | 60 | 53 | 76 | - | - | - | - | - | 20 | - |
| HST21-GYM | 99 | 98 | - | 84 | 91 | - | 86 | 22 | 50 | 99 | 99 |
| HST21-SPM | 97 | 95 | - | 79 | 96 | - | 100 | - | 53 | 99 | 99 |
| HST23-GYM | 93 | 92 | 100 | 97 | 71 | 77 | 100 | 100 | 49 | 99 | 99 |
| HST23-SPM | 98 | 97 | 97 | 99 | 97 | 76 | 100 | 100 | 49 | 99 | 99 |
| HST28-GYM | 97 | 98 | 100 | 100 | 22 | 45 | 37 | 94 | 27 | 100 | 100 |
| HST28-SPM | 100 | 100 | 100 | 100 | - | 39 | 29 | 89 | 25 | 97 | 100 |
| HST50-GYM | 96 | - | 90 | 97 | 92 | - | 41 | 29 | - | 43 | 38 |
| HST50-SPM | 93 | 41 | 97 | 98 | - | - | 57 | 30 | - | - | - |
| HST54-GYM | 96 | 95 | 96 | 78 | - | - | 48 | - | 39 | 53 | 24 |
| HST54-SPM | 95 | 92 | 93 | 71 | - | - | 47 | - | 35 | 52 | 25 |
| HST61-GYM | 73 | - | - | 76 | 63 | - | 59 | 59 | 22 | - | - |
| HST61-SPM | 81 | 41 | - | 62 | 68 | - | 43 | 62 | 26 | - | - |
| HST68-GYM | 93 | - | 90 | 97 | 91 | - | 30 | 21 | - | - | 21 |
| HST68-SPM | 38 | - | 50 | 24 | - | - | 45 | - | - | - | - |
| HST72-GYM | 95 | 97 | 98 | 35 | - | 28 | 60 | 100 | 47 | 97 | 69 |
| HST72-SPM | 97 | 97 | 99 | 56 | - | - | 50 | 96 | 28 | 86 | 59 |
| HST82-GYM | 100 | 97 | 90 | 92 | - | - | 41 | - | - | 50 | 40 |
| HST82-SPM | - | 47 | - | - | - | - | 26 | - | - | 39 | 38 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés-Albayay, C.; Silber, J.; Imhoff, J.F.; Asenjo, J.A.; Andrews, B.; Nouioui, I.; Dorador, C. The Polyextreme Ecosystem, Salar de Huasco at the Chilean Altiplano of the Atacama Desert Houses Diverse Streptomyces spp. with Promising Pharmaceutical Potentials. Diversity 2019, 11, 69. https://doi.org/10.3390/d11050069
Cortés-Albayay C, Silber J, Imhoff JF, Asenjo JA, Andrews B, Nouioui I, Dorador C. The Polyextreme Ecosystem, Salar de Huasco at the Chilean Altiplano of the Atacama Desert Houses Diverse Streptomyces spp. with Promising Pharmaceutical Potentials. Diversity. 2019; 11(5):69. https://doi.org/10.3390/d11050069
Chicago/Turabian StyleCortés-Albayay, Carlos, Johanna Silber, Johannes F. Imhoff, Juan A. Asenjo, Barbara Andrews, Imen Nouioui, and Cristina Dorador. 2019. "The Polyextreme Ecosystem, Salar de Huasco at the Chilean Altiplano of the Atacama Desert Houses Diverse Streptomyces spp. with Promising Pharmaceutical Potentials" Diversity 11, no. 5: 69. https://doi.org/10.3390/d11050069
APA StyleCortés-Albayay, C., Silber, J., Imhoff, J. F., Asenjo, J. A., Andrews, B., Nouioui, I., & Dorador, C. (2019). The Polyextreme Ecosystem, Salar de Huasco at the Chilean Altiplano of the Atacama Desert Houses Diverse Streptomyces spp. with Promising Pharmaceutical Potentials. Diversity, 11(5), 69. https://doi.org/10.3390/d11050069






