A Crosslinking Analysis of GAP-43 Interactions with Other Proteins in Differentiated N1E-115 Cells
Abstract
:1. Introduction
2. Results and Discussion
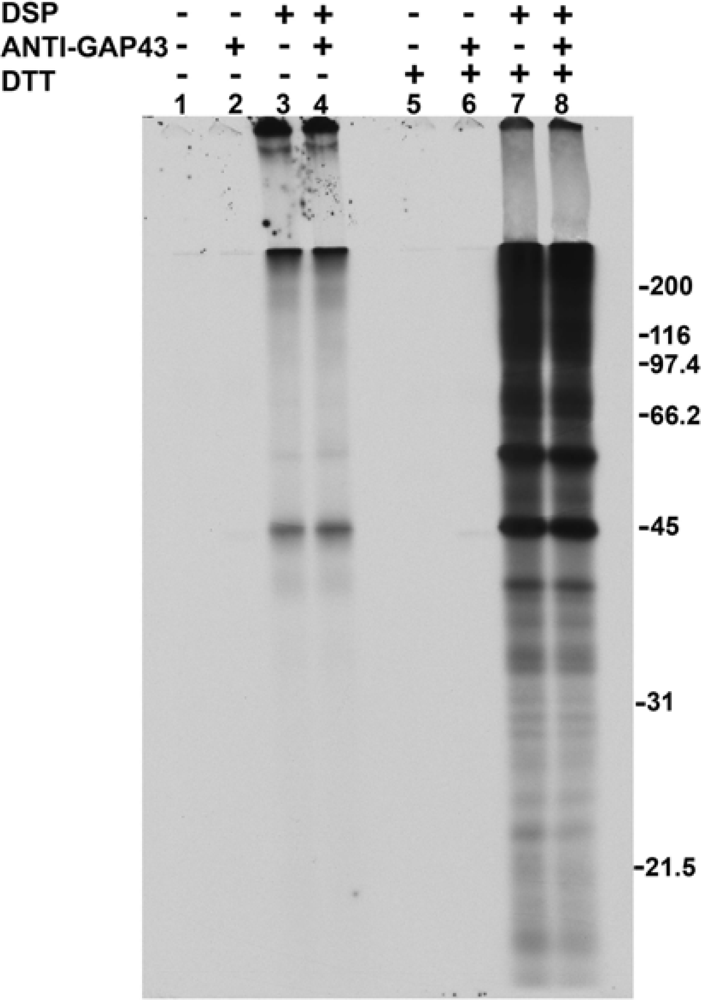
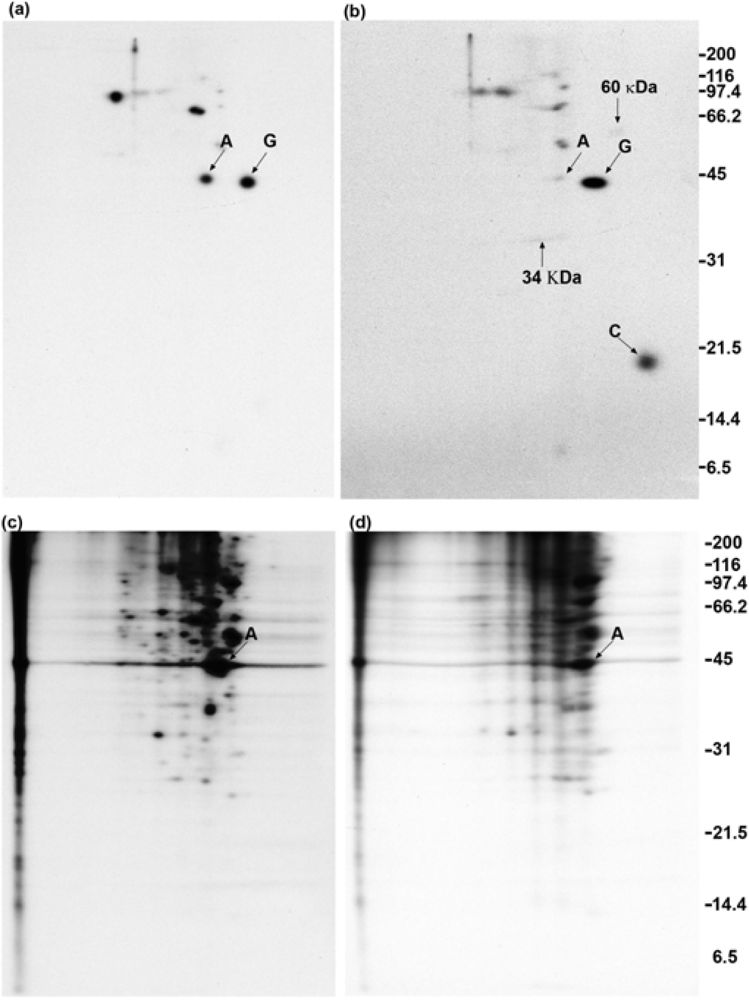
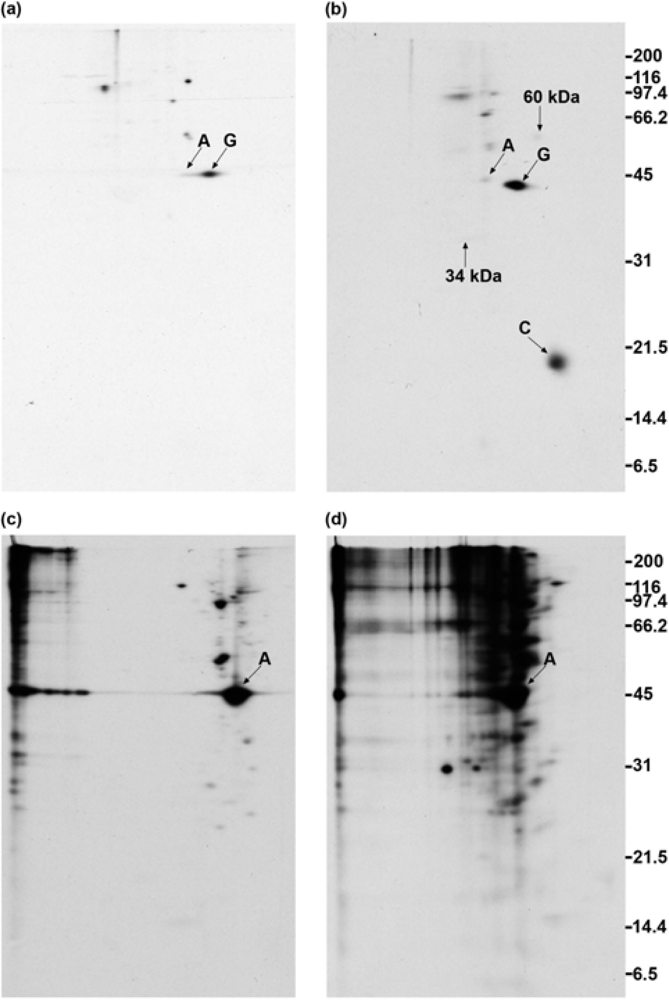
2.1. Addition of DSP to living cells yields crosslinked complexes that will not enter a one-dimensional gel
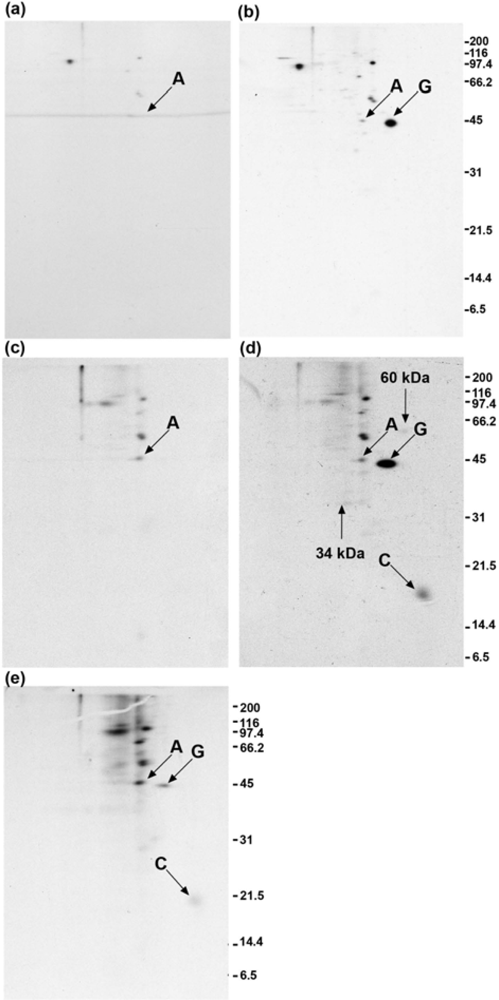
2.2. Calmodulin, but not actin or other proposed interaction partners, co-immunoprecipitates with GAP-43 after addition of crosslinker to living cells
2.3. GAP-43 does not appear in the 100,000 × g pellet fractions
2.4. Polyclonal anti-GAP-43 yields co-immunoprecipitated calmodulin but not actin following crosslinker treatment of cells
2.5. Quantitation of GAP-43 and calmodulin in immunoprecipitates
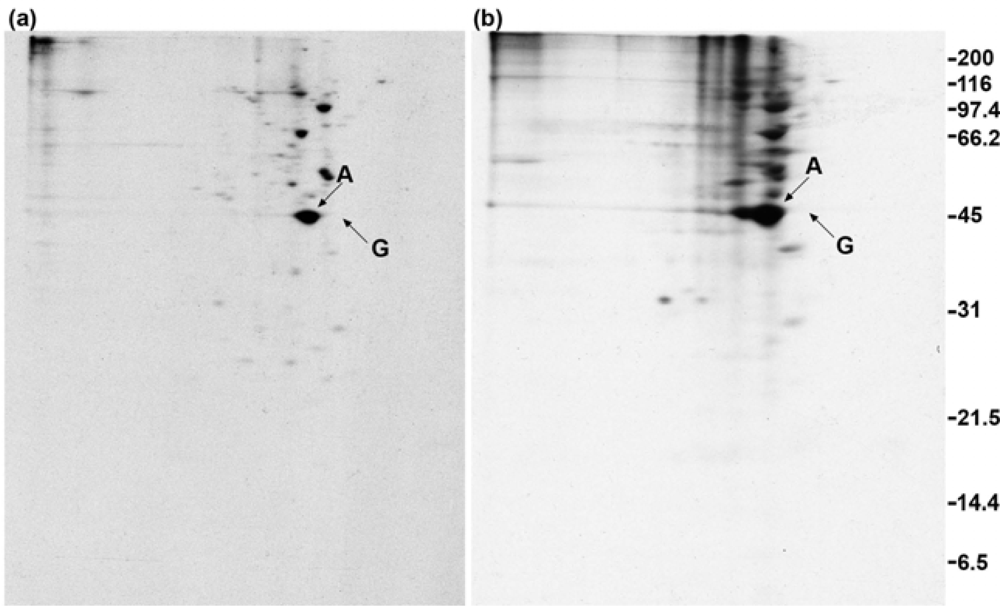
2.6. Anti-actin does not yield co-immunoprecipitated GAP-43 when cells are treated with crosslinker
2.7. GAP-43 does not sediment with the membrane skeleton when DMSO- or DSP-treated cells are lysed in nonionic detergent
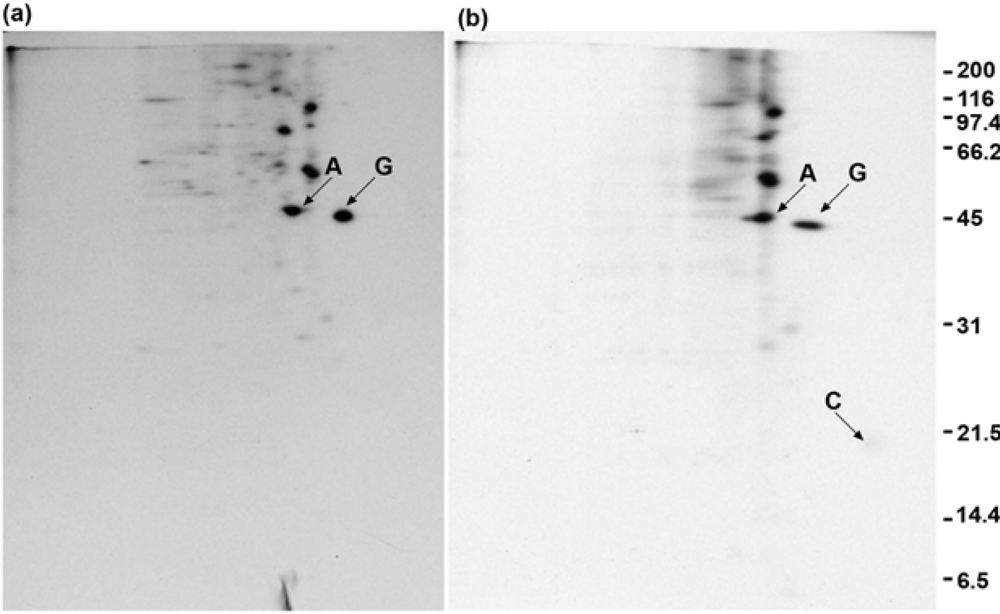
2.8. GAP-43 is crosslinked to calmodulin but not to actin if cells are lysed in nonionic detergent prior to the addition of crosslinker
3. Conclusions
4. Experimental Section
4.1. Materials
4.2. Cell culture and differentiation
4.3. Labeling of differentiated cells
4.4. Addition of crosslinker to living cells
4.5. Lysis of crosslinker-treated cells in SDS
4.6. Lysis of crosslinker-treated cells in nonionic detergent and isolation of the membrane skeleton
4.7. Addition of crosslinker to cell lysates
4.8. Immunoprecipitation of GAP-43, calmodulin, and actin
4.9. Gel electrophoresis
4.10. Determination of radioactivity in gels
Acknowledgments
References
- Benowitz, LI; Routtenberg, A. GAP-43: An intrinsic determinant of neuronal development and plasticity. Trends Neurosci 1997, 20, 84–91. [Google Scholar]
- Holahan, MR; Honegger, KS; Tabatadze, N; Routtenberg, A. GAP-43 gene expression regulates information storage. Learn. Mem 2007, 14, 407–415. [Google Scholar]
- Mosevitsky, MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol 2005, 245, 245–325. [Google Scholar]
- Denny, JB. Molecular mechanisms, biological actions, and neuropharmacology of the growth-associated protein GAP-43. Curr. Neuropharm 2006, 4, 293–304. [Google Scholar]
- Harel, NY; Strittmatter, SM. Can regenerating axons recapitulate developmental guidance during recovery from spinal cord injury? Nat. Rev. Neurosci 2006, 7, 603–616. [Google Scholar]
- Strittmatter, SM; Fankhauser, C; Huang, PL; Mashimo, H; Fishman, MC. Neuronal pathfinding is abnormal in mice lacking the neuronal growth cone protein GAP-43. Cell 1995, 80, 445–452. [Google Scholar]
- Denny, JB. Growth-associated protein of 43 kDa (GAP-43) is cleaved nonprocessively by the 20S proteasome. Eur. J. Biochem 2004, 271, 2480–2493. [Google Scholar]
- Skene, JHP; Virag, I. Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J. Cell Biol 1989, 108, 613–624. [Google Scholar]
- McLaughlin, RE; Denny, JB. Palmitoylation of GAP-43 by the ER-Golgi intermediate compartment and Golgi apparatus. Biochim. Biophys. Acta 1999, 1451, 82–92. [Google Scholar]
- Liang, X; Lu, Y; Neubert, TA; Resh, MD. Mass spectrometric analysis of GAP-43/neuromodulin reveals the presence of a variety of fatty acylated species. J. Biol. Chem 2002, 277, 33032–33040. [Google Scholar]
- Gauthier-Campbell, C; Bredt, DS; Murphy, TH; El-Husseini, AE. Regulation of dendritic branching and filopodial formation in hippocampal neurons by specific acylated protein motifs. Mol. Biol. Cell 2004, 15, 2205–2217. [Google Scholar]
- Patterson, SI; Skene, JH. A shift in protein S-palmitoylation, with persistence of growth-associated substrates, marks a critical period for synaptic plasticity in developing brain. J. Neurobiol 1999, 39, 423–437. [Google Scholar]
- Meiri, KF; Saffell, JL; Walsh, FS; Doherty, P. Neurite outgrowth stimulated by neural cell adhesion molecules requires growth-associated protein-43 (GAP-43) function and is associated with GAP-43 phosphorylation in growth cones. J. Neurosci 1998, 18, 10429–10437. [Google Scholar]
- Lalli, G; Hall, A. Ral GTPases regulate neurite branching through GAP-43 and the exocyst complex. J. Cell Biol 2005, 171, 857–869. [Google Scholar]
- Hens, JJH; Benfenati, F; Nielander, HB; Valtorta, F; Gispen, WH; De Graan, PNE. B-50/GAP-43 binds to actin filaments without affecting actin polymerization and filament organization. J. Neurochem 1993, 61, 1530–1533. [Google Scholar]
- He, Q; Dent, EW; Meiri, KF. Modulation of actin filament behavior by GAP-43 (neuromodulin) is dependent on the phosphorylation status of serine 41, the protein kinase C site. J. Neurosci 1997, 17, 3515–3524. [Google Scholar]
- Strittmatter, SM; Vartanian, T; Fishman, MC. GAP-43 as a plasticity protein in neuronal form and repair. J. Neurobiol 1992, 23, 507–520. [Google Scholar]
- Riederer, BM; Routtenberg, A. Can GAP-43 interact with brain spectrin? Mol. Brain Res 1999, 71, 345–348. [Google Scholar]
- Strittmatter, SM; Valenzuela, D; Kennedy, TE; Neer, EJ; Fishman, MC. Go is a major growth cone protein subject to regulation by GAP-43. Nature 1990, 344, 836–841. [Google Scholar]
- Haruta, T; Takami, N; Ohmura, M; Misumi, Y; Ikehara, Y. Ca2+-dependent interaction of the growth-associated protein GAP-43 with the synaptic core complex. Biochem. J 1997, 325, 455–463. [Google Scholar]
- Neve, RL; Coopersmith, R; McPhie, DL; Santeufemio, C; Pratt, KG; Murphy, CJ; Lynn, SD. The neuronal growth-associated protein GAP-43 interacts with rabaptin-5 and participates in endocytosis. J. Neurosci 1998, 18, 7757–7767. [Google Scholar]
- Chao, S; Benowitz, LI; Krainc, D; Irwin, N. Use of a two-hybrid system to investigate molecular interactions of GAP-43. Mol. Brain Res 1996, 40, 195–202. [Google Scholar]
- Chapman, ER; Au, D; Alexander, KA; Nicolson, TA; Storm, DR. Characterization of the calmodulin binding domain of neuromodulin. Functional significance of serine 41 and phenylalanine 42. J. Biol. Chem 1991, 266, 207–213. [Google Scholar]
- de Graan, PNE; Oestreicher, AB; de Wit, M; Kroef, M; Schrama, LH; Gispen, WH. Evidence for the binding of calmodulin to endogenous B-50 (GAP-43) in native synaptosomal plasma membranes. J. Neurochem 1990, 55, 2139–2141. [Google Scholar]
- Gamby, C; Waage, MC; Allen, RG; Baizer, L. Analysis of the role of calmodulin binding and sequestration in neuromodulin (GAP-43) function. J. Biol. Chem 1996, 271, 26698–26705. [Google Scholar]
- Laux, T; Fukami, K; Thelen, M; Golub, T; Frey, D; Caroni, P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P2 at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol 2000, 149, 1455–1471. [Google Scholar]
- Tong, J; Nguyen, L; Vidal, A; Simon, SA; Skene, JH Pate; McIntosh, TJ. Role of GAP-43 in sequestering phosphatidylinositol 4,5-bisphosphate to raft bilayers. Biophys. J 2008, 94, 125–133. [Google Scholar]
- Yuyama, K; Sekino-Suzuki, N; Sanai, Y; Kasahara, K. Translocation of activated heterotrimeric G protein Gαo to ganglioside-enriched detergent-resistant membrane rafts in developing cerebellum. J. Biol. Chem 2007, 282, 26392–26400. [Google Scholar]
- Brabet, P; Pantaloni, C; Bockaert, J; Homburger, V. Metabolism of two Go alpha isoforms in neuronal cells during differentiation. J. Biol. Chem 1991, 266, 12825–12828. [Google Scholar]
- Blikstad, I; Lazarides, E. Synthesis of spectrin in avian erythroid cells: Association of nascent polypeptide chains with the cytoskeleton. Proc. Natl. Acad. Sci. USA 1983, 80, 2637–2641. [Google Scholar]
- Nielsen, E; Christoforidis, S; Uttenweiler-Joseph, S; Miaczynska, M; Dewitte, F; Wilm, M; Hoflack, B; Zerial, M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol 2000, 151, 601–612. [Google Scholar]
- Gonzalo, S; Linder, ME. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol. Biol. Cell 1998, 9, 585–597. [Google Scholar]
- McQuarrie, IG; Brady, ST; Lasek, RJ. Diversity in the axonal transport of structural proteins: Major differences between optic and spinal axons in the rat. J. Neurosci 1986, 6, 1593–1605. [Google Scholar]
- Moss, DJ; Fernyhough, P; Chapman, K; Baizer, L; Bray, D; Allsopp, T. Chicken growth-associated protein GAP-43 is tightly bound to the actin-rich neuronal membrane skeleton. J. Neurochem 1990, 54, 729–736. [Google Scholar]
- Meiri, KF; Gordon-Weeks, PR. GAP-43 in growth cones is associated with areas of membrane that are tightly bound to substrate and is a component of a membrane skeleton subcellular fraction. J. Neurosci 1990, 10, 256–266. [Google Scholar]
- Nebl, T; Pestonjamasp, KN; Leszyk, JD; Crowley, JL; Oh, SW; Luna, EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J. Biol. Chem 2002, 277, 43399–43409. [Google Scholar]
- Faix, J; Rottner, K. The making of filopodia. Curr. Opin. Cell Biol 2006, 18, 18–25. [Google Scholar]
- Aarts, LHJ; Verkade, P; Schrama, LH; Oestreicher, AB; Gispen, WH; Schotman, P. Local accumulation of B-50/GAP-43 evoke excessive bleb formation in PC12 cells. Mol. Neurobiol 1999, 20, 17–28. [Google Scholar]
- Mattila, PK; Pykalainen, A; Saarikangas, J; Paavilainen, VO; Vihinen, H; Jokitalo, E; Lappalainen, P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol 2007, 176, 953–964. [Google Scholar]
- Tapp, H; Al-Naggar, IM; Yarmola, EG; Harrison, A; Shaw, G; Edison, AS; Bubb, MR. MARCKS is a natively unfolded protein with an inaccessible actin-binding site. Evidence for long-range intramolecular interactions. J. Biol. Chem 2005, 280, 9946–9956. [Google Scholar]
- Apel, ED; Litchfield, DW; Clark, RH; Krebs, EG; Storm, DR. Phosphorylation of neuromodulin (GAP-43) by casein kinase II. Identification of phosphorylation sites and regulation by calmodulin. J. Biol. Chem 1991, 266, 10544–10551. [Google Scholar]
- Liu, Y; Fisher, DA; Storm, DR. Intracellular sorting of neuromodulin (GAP-43) mutants modified in the membrane targeting domain. J. Neurosci 1994, 14, 5807–5817. [Google Scholar]
- Meiri, KF; Bickerstaff, LE; Schwob, JE. Monoclonal antibodies show that kinase C phosphorylation of GAP-43 during axonogenesis is both spatially and temporally restricted in vivo. J. Cell Biol 1991, 112, 991–1005. [Google Scholar]
- Baker, LP; Storm, DR. Dynamic palmitoylation of neuromodulin (GAP-43) in cultured rat cerebellar neurons and mouse N1E-115 cells. Neurosci. Lett 1997, 234, 156–160. [Google Scholar]
- Carneiro, AMD; Blakely, RD. Serotonin-, protein kinase C-, and Hic-5-associated redistribution of the platelet serotonin transporter. J. Biol. Chem 2006, 281, 24769–24780. [Google Scholar]
- Anderson, L; Denny, JB. Protein translocation in the endoplasmic reticulum. Ultraviolet light induces the noncovalent association of nascent peptides with translocon proteins. J. Biol. Chem 1992, 267, 23916–23921. [Google Scholar]
- O’Farrell, PH. High-resolution two-dimensional electrophoresis of proteins. J. Biol. Chem 1975, 250, 4007–4021. [Google Scholar]

| GAP43 | CaM | % | |
|---|---|---|---|
| Figure 2b DMSO | 2967 | 0 | 0 |
| Figure 2d DSP | 1933 | 278 | 14 |
| Figure 2e antiCaM | 322 | ||
| Figure 4a DMSO | 791 | ||
| Figure 4b DSP | 619 | ||
| Figure 7b DSP | 1650 | 643 | 39 |
© 2008 by MDPI This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ollom, C.M.; Denny, J.B. A Crosslinking Analysis of GAP-43 Interactions with Other Proteins in Differentiated N1E-115 Cells. Int. J. Mol. Sci. 2008, 9, 1753-1771. https://doi.org/10.3390/ijms9091753
Ollom CM, Denny JB. A Crosslinking Analysis of GAP-43 Interactions with Other Proteins in Differentiated N1E-115 Cells. International Journal of Molecular Sciences. 2008; 9(9):1753-1771. https://doi.org/10.3390/ijms9091753
Chicago/Turabian StyleOllom, Callise M., and John B. Denny. 2008. "A Crosslinking Analysis of GAP-43 Interactions with Other Proteins in Differentiated N1E-115 Cells" International Journal of Molecular Sciences 9, no. 9: 1753-1771. https://doi.org/10.3390/ijms9091753




