Sublethal RNA Oxidation as a Mechanism for Neurodegenerative Disease
Abstract
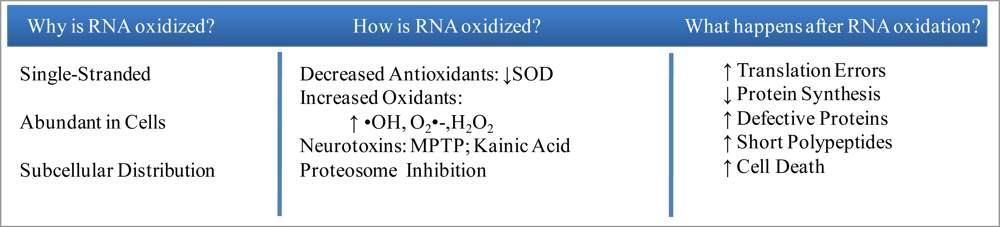
:1. Introduction
2. Significance of RNA Studies in Neurodegenerative Disease Research
3. RNA Oxidation in Various Neurological Diseases
4. Experimental Models
5. Time Course of RNA Oxidation
6. Types of Reactive Oxygen Species and Relevance to RNA
7. Pathogenic Cascade Initiated by RNA Oxidation
8. Repair Mechanisms
9. Conclusions
Acknowledgments
References and Notes
- Hirtz, D; Thurman, DJ; Gwinn-Hardy, K; Mohamed, M; Chaudhuri, AR; Zalutsky, R. How common are the “common” neurologic disorders? Neurology 2007, 68, 326–337. [Google Scholar] [Green Version]
- Sayre, LM; Smith, MA; Perry, G. Chemistry and biochemistry of oxidative stress in neurodegenerative disease. Curr. Med. Chem. 2001, 8, 721–738. [Google Scholar] [Green Version]
- Ischiropoulos, H; Beckman, JS. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J. Clin. Invest. 2003, 111, 163–169. [Google Scholar] [Green Version]
- Jenner, P. Oxidative stress in Parkinson's disease. Ann. Neurol. 2003, 53 Suppl 3, S26–36, discussion S36–28. [Google Scholar] [Green Version]
- Andersen, JK. Oxidative stress in neurodegeneration: cause or consequence? Nat. Med. 2004, 10(Suppl), S18–25. [Google Scholar] [Green Version]
- Barnham, KJ; Masters, CL; Bush, AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov 2004, 3, 205–214. [Google Scholar] [Green Version]
- Barber, SC; Mead, RJ; Shaw, PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim. Biophys. Acta 2006, 1762, 1051–1067. [Google Scholar] [Green Version]
- Lin, MT; Beal, MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [Green Version]
- Nunomura, A; Castellani, RJ; Zhu, X; Moreira, PI; Perry, G; Smith, MA. Involvement of oxidative stress in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2006, 65, 631–641. [Google Scholar] [Green Version]
- Nunomura, A; Perry, G; Pappolla, MA; Wade, R; Hirai, K; Chiba, S; Smith, MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J. Neurosci. 1999, 19, 1959–1964. [Google Scholar] [Green Version]
- Li, Z; Wu, J; Deleo, CJ. RNA damage and surveillance under oxidative stress. IUBMB life 2006, 58, 581–588. [Google Scholar] [Green Version]
- Bregeon, D; Sarasin, A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat. Res. 2005, 577, 293–302. [Google Scholar] [Green Version]
- Moreira, PI; Nunomura, A; Nakamura, M; Takeda, A; Shenk, JC; Aliev, G; Smith, MA; Perry, G. Nucleic acid oxidation in Alzheimer disease. Free Radic. Biol. Med. 2008, 44, 1493–1505. [Google Scholar] [Green Version]
- Hofer, T; Seo, AY; Prudencio, M; Leeuwenburgh, C. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biol. Chem. 2006, 387, 103–111. [Google Scholar] [Green Version]
- Shen, Z; Wu, W; Hazen, SL. Activated leukocytes oxidatively damage DNA, RNA, and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry (Mosc). 2000, 39, 5474–5482. [Google Scholar] [Green Version]
- Wamer, WG; Wei, RR. In vitro photooxidation of nucleic acids by ultraviolet A radiation. Photochem. Photobiol. 1997, 65, 560–563. [Google Scholar] [Green Version]
- Hofer, T; Badouard, C; Bajak, E; Ravanat, JL; Mattsson, A; Cotgreave, IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005, 386, 333–337. [Google Scholar] [Green Version]
- Fiala, ES; Conaway, CC; Mathis, JE. Oxidative DNA and RNA damage in the livers of Sprague-Dawley rats treated with the hepatocarcinogen 2-nitropropane. Cancer Res. 1989, 49, 5518–5522. [Google Scholar] [Green Version]
- Weimann, A; Belling, D; Poulsen, HE. Quantification of 8-oxo-guanine and guanine as the nucleobase, nucleoside and deoxynucleoside forms in human urine by high-performance liquid chromatography-electrospray tandem mass spectrometry. Nucleic Acids Res 2002, 30, E7. [Google Scholar] [Green Version]
- Szymanski, M; Barciszewska, MZ; Erdmann, VA; Barciszewski, J. A new frontier for molecular medicine: noncoding RNAs. Biochim. Biophys. Acta 2005, 1756, 65–75. [Google Scholar] [Green Version]
- Costa, FF. Non-coding RNAs: new players in eukaryotic biology. Gene 2005, 357, 83–94. [Google Scholar] [Green Version]
- Cao, X; Yeo, G; Muotri, AR; Kuwabara, T; Gage, FH. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006, 29, 77–103. [Google Scholar] [Green Version]
- Mehler, MF; Mattick, JS. Non-coding RNAs in the nervous system. J. Physiol. 2006, 575, 333–341. [Google Scholar] [Green Version]
- Taft, RJ; Pheasant, M; Mattick, JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 2007, 29, 288–299. [Google Scholar] [Green Version]
- Perkins, DO; Jeffries, C; Sullivan, P. Expanding the ‘central dogma’: the regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Mol. Psychiatry 2005, 10, 69–78. [Google Scholar] [Green Version]
- Taylor, JP; Hardy, J; Fischbeck, KH. Toxic proteins in neurodegenerative disease. Science 2002, 296, 1991–1995. [Google Scholar] [Green Version]
- van Leeuwen, FW; de Kleijn, DP; van den Hurk, HH; Neubauer, A; Sonnemans, MA; Sluijs, JA; Koycu, S; Ramdjielal, RD; Salehi, A; Martens, GJ; Grosveld, FG; Peter, J; Burbach, H; Hol, EM. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science 1998, 279, 242–247. [Google Scholar] [Green Version]
- Lee, JW; Beebe, K; Nangle, LA; Jang, J; Longo-Guess, CM; Cook, SA; Davisson, MT; Sundberg, JP; Schimmel, P; Ackerman, SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 2006, 443, 50–55. [Google Scholar] [Green Version]
- Evans, MD; Dizdaroglu, M; Cooke, MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [Green Version]
- Cooke, MS; Olinski, R; Evans, MD. Does measurement of oxidative damage to DNA have clinical significance? Clin. Chim. Acta 2006, 365, 30–49. [Google Scholar] [Green Version]
- Kasai, H; Crain, PF; Kuchino, Y; Nishimura, S; Ootsuyama, A; Tanooka, H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 1986, 7, 1849–1851. [Google Scholar] [Green Version]
- Ames, BN; Gold, LS. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 1991, 250, 3–16. [Google Scholar] [Green Version]
- Yanagawa, H; Ogawa, Y; Ueno, M. Redox ribonucleosides. Isolation and characterization of 5-hydroxyuridine, 8-hydroxyguanosine, and 8-hydroxyadenosine from Torula yeast RNA. J. Biol. Chem. 1992, 267, 13320–13326. [Google Scholar] [Green Version]
- Schneider, JE, Jr; Phillips, JR; Pye, Q; Maidt, ML; Price, S; Floyd, RA. Methylene blue and rose bengal photoinactivation of RNA bacteriophages: comparative studies of 8-oxoguanine formation in isolated RNA. Arch. Biochem. Biophys. 1993, 301, 91–97. [Google Scholar] [Green Version]
- Rhee, Y; Valentine, MR; Termini, J. Oxidative base damage in RNA detected by reverse transcriptase. Nucleic Acids Res 1995, 23, 3275–3282. [Google Scholar] [Green Version]
- Barciszewski, J; Barciszewska, MZ; Siboska, G; Rattan, SI; Clark, BF. Some unusual nucleic acid bases are products of hydroxyl radical oxidation of DNA and RNA. Mol. Biol. Rep. 1999, 26, 231–238. [Google Scholar] [Green Version]
- Yin, B; Whyatt, RM; Perera, FP; Randall, MC; Cooper, TB; Santella, RM. Determination of 8-hydroxydeoxyguanosine by an immunoaffinity chromatography-monoclonal antibody-based ELISA. Free Radic. Biol. Med. 1995, 18, 1023–1032. [Google Scholar] [Green Version]
- Park, EM; Shigenaga, MK; Degan, P; Korn, TS; Kitzler, JW; Wehr, CM; Kolachana, P; Ames, BN. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivatives from biological fluids with a monoclonal antibody column. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 3375–3379. [Google Scholar] [Green Version]
- Zhang, J; Perry, G; Smith, MA; Robertson, D; Olson, SJ; Graham, DG; Montine, TJ. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am. J. Pathol. 1999, 154, 1423–1429. [Google Scholar] [Green Version]
- Nunomura, A; Perry, G; Aliev, G; Hirai, K; Takeda, A; Balraj, EK; Jones, PK; Ghanbari, H; Wataya, T; Shimohama, S; Chiba, S; Atwood, CS; Petersen, RB; Smith, MA. Oxidative damage is the earliest event in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2001, 60, 759–767. [Google Scholar] [Green Version]
- Nunomura, A; Perry, G; Pappolla, MA; Friedland, RP; Hirai, K; Chiba, S; Smith, MA. Neuronal oxidative stress precedes amyloid-beta deposition in Down syndrome. J. Neuropathol. Exp. Neurol. 2000, 59, 1011–1017. [Google Scholar] [Green Version]
- Nunomura, A; Chiba, S; Kosaka, K; Takeda, A; Castellani, RJ; Smith, MA; Perry, G. Neuronal RNA oxidation is a prominent feature of dementia with Lewy bodies. Neuroreport 2002, 13, 2035–2039. [Google Scholar] [Green Version]
- Guentchev, M; Siedlak, SL; Jarius, C; Tagliavini, F; Castellani, RJ; Perry, G; Smith, MA; Budka, H. Oxidative damage to nucleic acids in human prion disease. Neurobiol. Dis. 2002, 9, 275–281. [Google Scholar] [Green Version]
- Hayashi, M; Arai, N; Satoh, J; Suzuki, H; Katayama, K; Tamagawa, K; Morimatsu, Y. Neurodegenerative mechanisms in subacute sclerosing panencephalitis. J. Child Neurol. 2002, 17, 725–730. [Google Scholar] [Green Version]
- Nunomura, A; Chiba, S; Lippa, CF; Cras, P; Kalaria, RN; Takeda, A; Honda, K; Smith, MA; Perry, G. Neuronal RNA oxidation is a prominent feature of familial Alzheimer's disease. Neurobiol. Dis. 2004, 17, 108–113. [Google Scholar] [Green Version]
- Petersen, RB; Siedlak, SL; Lee, HG; Kim, YS; Nunomura, A; Tagliavini, F; Ghetti, B; Cras, P; Moreira, PI; Castellani, RJ; Guentchev, M; Budka, H; Ironside, JW; Gambetti, P; Smith, MA; Perry, G. Redox metals and oxidative abnormalities in human prion diseases. Acta Neuropathol 2005, 110, 232–238. [Google Scholar] [Green Version]
- Hayashi, M; Araki, S; Kohyama, J; Shioda, K; Fukatsu, R. Oxidative nucleotide damage and superoxide dismutase expression in the brains of xeroderma pigmentosum group A and Cockayne syndrome. Brain Dev. 2005, 27, 34–38. [Google Scholar] [Green Version]
- Tateyama, M; Takeda, A; Onodera, Y; Matsuzaki, M; Hasegawa, T; Nunomura, A; Hirai, K; Perry, G; Smith, MA; Itoyama, Y. Oxidative stress and predominant Abeta42(43) deposition in myopathies with rimmed vacuoles. Acta Neuropathol. (Berl). 2003, 105, 581–585. [Google Scholar] [Green Version]
- Martinet, W; de Meyer, GR; Herman, AG; Kockx, MM. Reactive oxygen species induce RNA damage in human atherosclerosis. Eur. J. Clin. Invest. 2004, 34, 323–327. [Google Scholar] [Green Version]
- Hofer, T; Marzetti, E; Xu, J; Seo, AY; Gulec, S; Knutson, MD; Leeuwenburgh, C; Dupont-Versteegden, EE. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Exp. Gerontol. 2008, in press. [Google Scholar]
- Shan, X. The identification and characterization of oxidized RNAs in Alzheimer's disease. J. Neurosci. 2003, 23, 4913–4921. [Google Scholar] [Green Version]
- Honda, K; Smith, MA; Zhu, X; Baus, D; Merrick, WC; Tartakoff, AM; Hattier, T; Harris, PL; Siedlak, SL; Fujioka, H; Liu, Q; Moreira, PI; Miller, FP; Nunomura, A; Shimohama, S; Perry, G. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. J. Biol. Chem. 2005, 280, 20978–20986. [Google Scholar] [Green Version]
- Ding, Q; Markesbery, WR; Chen, Q; Li, F; Keller, JN. Ribosome dysfunction is an early event in Alzheimer's disease. J. Neurosci. 2005, 25, 9171–9175. [Google Scholar] [Green Version]
- Ding, Q; Markesbery, WR; Cecarini, V; Keller, JN. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem. Res. 2006, 31, 705–710. [Google Scholar] [Green Version]
- Shan, X; Lin, CL. Quantification of oxidized RNAs in Alzheimer's disease. Neurobiol. Aging 2006, 27, 657–662. [Google Scholar] [Green Version]
- Abe, T; Tohgi, H; Isobe, C; Murata, T; Sato, C. Remarkable increase in the concentration of 8-hydroxyguanosine in cerebrospinal fluid from patients with Alzheimer's disease. J. Neurosci. Res. 2002, 70, 447–450. [Google Scholar] [Green Version]
- Kikuchi, A; Takeda, A; Onodera, H; Kimpara, T; Hisanaga, K; Sato, N; Nunomura, A; Castellani, RJ; Perry, G; Smith, MA; Itoyama, Y. Systemic increase of oxidative nucleic acid damage in Parkinson's disease and multiple system atrophy. Neurobiol. Dis. 2002, 9, 244–248. [Google Scholar] [Green Version]
- Abe, T; Isobe, C; Murata, T; Sato, C; Tohgi, H. Alteration of 8-hydroxyguanosine concentrations in the cerebrospinal fluid and serum from patients with Parkinson's disease. Neurosci. Lett. 2003, 336, 105–108. [Google Scholar] [Green Version]
- Liu, J; Head, E; Gharib, AM; Yuan, W; Ingersoll, RT; Hagen, TM; Cotman, CW; Ames, BN. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 2356–2361. [Google Scholar] [Green Version]
- Row, BW; Liu, R; Xu, W; Kheirandish, L; Gozal, D. Intermittent hypoxia is associated with oxidative stress and spatial learning deficits in the rat. Am. J. Respir. Crit. Care Med. 2003, 167, 1548–1553. [Google Scholar] [Green Version]
- Yamaguchi, H; Kajitani, K; Dan, Y; Furuichi, M; Ohno, M; Sakumi, K; Kang, D; Nakabeppu, Y. MTH1, an oxidized purine nucleoside triphosphatase, protects the dopamine neurons from oxidative damage in nucleic acids caused by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Cell Death Differ. 2006, 13, 551–563. [Google Scholar] [Green Version]
- Foksinski, M; Rozalski, R; Guz, J; Ruszkowska, B; Sztukowska, P; Piwowarski, M; Klungland, A; Olinski, R. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radic. Biol. Med. 2004, 37, 1449–1454. [Google Scholar] [Green Version]
- Javitch, JA; D'Amato, RJ; Strittmatter, SM; Snyder, SH. Parkinsonism-inducing neurotoxin, N-methyl-4-phenyl-1,2,3,6 -tetrahydropyridine: uptake of the metabolite N-methyl-4-phenylpyridine by dopamine neurons explains selective toxicity. Proc. Natl. Acad. Sci. U. S. A. 1985, 82, 2173–2177. [Google Scholar] [Green Version]
- Wang, Q; Yu, S; Simonyi, A; Sun, GY; Sun, AY. Kainic acid-mediated excitotoxicity as a model for neurodegeneration. Mol. Neurobiol. 2005, 31, 3–16. [Google Scholar] [Green Version]
- Kajitani, K; Yamaguchi, H; Dan, Y; Furuichi, M; Kang, D; Nakabeppu, Y. MTH1, an oxidized purine nucleoside triphosphatase, suppresses the accumulation of oxidative damage of nucleic acids in the hippocampal microglia during kainate-induced excitotoxicity. J. Neurosci. 2006, 26, 1688–1698. [Google Scholar] [Green Version]
- Rosen, DR; Siddique, T; Patterson, D; Figlewicz, DA; Sapp, P; Hentati, A; Donaldson, D; Goto, J; O'Regan, JP; Deng, HX; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [Green Version]
- Gurney, ME; Pu, H; Chiu, AY; Dal Canto, MC; Polchow, CY; Alexander, DD; Caliendo, J; Hentati, A; Kwon, YW; Deng, HX; et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science 1994, 264, 1772–1775. [Google Scholar] [Green Version]
- Chang, Y; Shan, X; Lin, CL. RNA oxidation is an early event preceding motor neuron death in ALS. Soc. Neurosci. Abstr. 2004, 96.93. [Google Scholar]
- Ding, Q; Dimayuga, E; Markesbery, WR; Keller, JN. Proteasome inhibition increases DNA and RNA oxidation in astrocyte and neuron cultures. J. Neurochem. 2004, 91, 1211–1218. [Google Scholar] [Green Version]
- Shan, X; Chang, Y; Lin, CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007, 21, 2753–2764. [Google Scholar] [Green Version]
- Ding, Q; Cecarini, V; Keller, JN. Interplay between protein synthesis and degradation in the CNS: physiological and pathological implications. Trends Neurosci. 2007, 30, 31–36. [Google Scholar] [Green Version]
- Keller, JN; Schmitt, FA; Scheff, SW; Ding, Q; Chen, Q; Butterfield, DA; Markesbery, WR. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology 2005, 64, 1152–1156. [Google Scholar] [Green Version]
- Butterfield, DA; Reed, TT; Perluigi, M; De Marco, C; Coccia, R; Keller, JN; Markesbery, WR; Sultana, R. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: implications for the role of nitration in the progression of Alzheimer's disease. Brain Res. 2007, 1148, 243–248. [Google Scholar] [Green Version]
- Pratico, D; Clark, CM; Liun, F; Rokach, J; Lee, VY; Trojanowski, JQ. Increase of brain oxidative stress in mild cognitive impairment: a possible predictor of Alzheimer disease. Arch. Neurol. 2002, 59, 972–976. [Google Scholar] [Green Version]
- Migliore, L; Fontana, I; Trippi, F; Colognato, R; Coppede, F; Tognoni, G; Nucciarone, B; Siciliano, G. Oxidative DNA damage in peripheral leukocytes of mild cognitive impairment and AD patients. Neurobiol. Aging 2005, 26, 567–573. [Google Scholar] [Green Version]
- Rinaldi, P; Polidori, MC; Metastasio, A; Mariani, E; Mattioli, P; Cherubini, A; Catani, M; Cecchetti, R; Senin, U; Mecocci, P. Plasma antioxidants are similarly depleted in mild cognitive impairment and in Alzheimer's disease. Neurobiol. Aging 2003, 24, 915–919. [Google Scholar] [Green Version]
- Guidi, I; Galimberti, D; Lonati, S; Novembrino, C; Bamonti, F; Tiriticco, M; Fenoglio, C; Venturelli, E; Baron, P; Bresolin, N; Scarpini, E. Oxidative imbalance in patients with mild cognitive impairment and Alzheimer's disease. Neurobiol. Aging 2006, 27, 262–269. [Google Scholar] [Green Version]
- Moreira, PI; Zhu, X; Nunomura, A; Smith, MA; Perry, G. Therapeutic options in Alzheimer's disease. Expert Rev. Neurother. 2006, 6, 897–910. [Google Scholar] [Green Version]
- Liu, Q; Xie, F; Rolston, R; Moreira, PI; Nunomura, A; Zhu, X; Smith, MA; Perry, G. Prevention and treatment of Alzheimer disease and aging: antioxidants. Mini Rev. Med. Chem. 2007, 7, 171–180. [Google Scholar] [Green Version]
- Sayre, LM; Perry, G; Smith, MA. In situ methods for detection and localization of markers of oxidative stress: application in neurodegenerative disorders. Methods Enzymol. 1999, 309, 133–152. [Google Scholar] [Green Version]
- Smith, MA; Perry, G; Richey, PL; Sayre, LM; Anderson, VE; Beal, MF; Kowall, N. Oxidative damage in Alzheimer's. Nature 1996, 382, 120–121. [Google Scholar] [Green Version]
- Sayre, LM; Zelasko, DA; Harris, PL; Perry, G; Salomon, RG; Smith, MA. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J. Neurochem. 1997, 68, 2092–2097. [Google Scholar] [Green Version]
- Casadesus, G; Smith, MA; Basu, S; Hua, J; Capobianco, DE; Siedlak, SL; Zhu, X; Perry, G. Increased isoprostane and prostaglandin are prominent in neurons in Alzheimer disease. Mol. Neurodegener. 2007, 2, 2. [Google Scholar] [Green Version]
- Castellani, RJ; Harris, PL; Sayre, LM; Fujii, J; Taniguchi, N; Vitek, MP; Founds, H; Atwood, CS; Perry, G; Smith, MA. Active glycation in neurofibrillary pathology of Alzheimer disease: N(epsilon)-(carboxymethyl) lysine and hexitol-lysine. Free Radic. Biol. Med. 2001, 31, 175–180. [Google Scholar] [Green Version]
- Coyle, JT; Puttfarcken, P. Oxidative stress, glutamate, and neurodegenerative disorders. Science 1993, 262, 689–695. [Google Scholar] [Green Version]
- Mattson, MP; Chan, SL; Duan, W. Modification of brain aging and neurodegenerative disorders by genes, diet, and behavior. Physiol. Rev. 2002, 82, 637–672. [Google Scholar] [Green Version]
- Halliwell, B. Reactive oxygen species and the central nervous system. J. Neurochem. 1992, 59, 1609–1623. [Google Scholar] [Green Version]
- Joenje, H. Genetic toxicology of oxygen. Mutat. Res. 1989, 219, 193–208. [Google Scholar] [Green Version]
- Takahashi, MA; Asada, K. Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch. Biochem. Biophys. 1983, 226, 558–566. [Google Scholar] [Green Version]
- Schubert, J; Wilmer, JW. Does hydrogen peroxide exist "free" in biological systems? Free Radic. Biol. Med. 1991, 11, 545–555. [Google Scholar] [Green Version]
- Hirai, K; Aliev, G; Nunomura, A; Fujioka, H; Russell, RL; Atwood, CS; Johnson, AB; Kress, Y; Vinters, HV; Tabaton, M; Shimohama, S; Cash, AD; Siedlak, SL; Harris, PL; Jones, PK; Petersen, RB; Perry, G; Smith, MA. Mitochondrial abnormalities in Alzheimer's disease. J. Neurosci. 2001, 21, 3017–3023. [Google Scholar] [Green Version]
- Perry, G; Nunomura, A; Cash, AD; Taddeo, MA; Hirai, K; Aliev, G; Avila, J; Wataya, T; Shimohama, S; Atwood, CS; Smith, MA. Reactive oxygen: its sources and significance in Alzheimer disease. J. Neural Transm. Suppl. 2002, 69–75. [Google Scholar]
- Perry, G; Nunomura, A; Hirai, K; Zhu, X; Perez, M; Avila, J; Castellani, RJ; Atwood, CS; Aliev, G; Sayre, LM; Takeda, A; Smith, MA. Is oxidative damage the fundamental pathogenic mechanism of Alzheimer's and other neurodegenerative diseases? Free Radic. Biol. Med. 2002, 33, 1475–1479. [Google Scholar] [Green Version]
- Smith, MA. Metabolic, metallic, and mitotic sources of oxidative stress in Alzheimer disease. Antioxid. Redox Signal. 2000, 2, 413–420. [Google Scholar] [Green Version]
- Gu, G; Reyes, PE; Golden, GT; Woltjer, RL; Hulette, C; Montine, TJ; Zhang, J. Mitochondrial DNA deletions/rearrangements in parkinson disease and related neurodegenerative disorders. J. Neuropathol. Exp. Neurol. 2002, 61, 634–639. [Google Scholar] [Green Version]
- Schapira, AH; Cooper, JM; Dexter, D; Clark, JB; Jenner, P; Marsden, CD. Mitochondrial complex I deficiency in Parkinson's disease. J. Neurochem. 1990, 54, 823–827. [Google Scholar] [Green Version]
- Sofic, E; Riederer, P; Heinsen, H; Beckmann, H; Reynolds, GP; Hebenstreit, G; Youdim, MB. Increased iron (III) and total iron content in post mortem substantia nigra of parkinsonian brain. J. Neural Transm. 1988, 74, 199–205. [Google Scholar] [Green Version]
- Berg, D; Roggendorf, W; Schroder, U; Klein, R; Tatschner, T; Benz, P; Tucha, O; Preier, M; Lange, KW; Reiners, K; Gerlach, M; Becker, G. Echogenicity of the substantia nigra: association with increased iron content and marker for susceptibility to nigrostriatal injury. Arch. Neurol. 2002, 59, 999–1005. [Google Scholar] [Green Version]
- Ishibashi, T; Hayakawa, H; Ito, R; Miyazawa, M; Yamagata, Y; Sekiguchi, M. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res 2005, 33, 3779–3784. [Google Scholar] [Green Version]
- Taddei, F; Hayakawa, H; Bouton, M; Cirinesi, A; Matic, I; Sekiguchi, M; Radman, M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 1997, 278, 128–130. [Google Scholar] [Green Version]
- Tanaka, M; Chock, PB; Stadtman, ER. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 66–71. [Google Scholar] [Green Version]
- Gong, X; Tao, R; Li, Z. Quantification of RNA damage by reverse transcription polymerase chain reactions. Anal. Biochem. 2006, 357, 58–67. [Google Scholar] [Green Version]
- Bellacosa, A; Moss, EG. RNA repair: damage control. Curr. Biol. 2003, 13, R482–484. [Google Scholar] [Green Version]
- Deutscher, MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 2006, 34, 659–666. [Google Scholar] [Green Version]
- Sheth, U; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [Green Version]
- Sweet, TJ; Boyer, B; Hu, W; Baker, KE; Coller, J. Microtubule disruption stimulates P-body formation. RNA 2007, 13, 493–502. [Google Scholar] [Green Version]
- Kedersha, N; Stoecklin, G; Ayodele, M; Yacono, P; Lykke-Andersen, J; Fritzler, MJ; Scheuner, D; Kaufman, RJ; Golan, DE; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [Green Version]
- Aas, PA; Otterlei, M; Falnes, PO; Vagbo, CB; Skorpen, F; Akbari, M; Sundheim, O; Bjoras, M; Slupphaug, G; Seeberg, E; Krokan, HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar] [Green Version]
- Krokan, HE; Kavli, B; Slupphaug, G. Novel aspects of macromolecular repair and relationship to human disease. J. Mol. Med. 2004, 82, 280–297. [Google Scholar] [Green Version]
- Nakabeppu, Y; Tsuchimoto, D; Ichinoe, A; Ohno, M; Ide, Y; Hirano, S; Yoshimura, D; Tominaga, Y; Furuichi, M; Sakumi, K. Biological significance of the defense mechanisms against oxidative damage in nucleic acids caused by reactive oxygen species: from mitochondria to nuclei. Ann. N. Y. Acad. Sci. 2004, 1011, 101–111. [Google Scholar] [Green Version]
- Hayakawa, H; Hofer, A; Thelander, L; Kitajima, S; Cai, Y; Oshiro, S; Yakushiji, H; Nakabeppu, Y; Kuwano, M; Sekiguchi, M. Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry (Mosc). 1999, 38, 3610–3614. [Google Scholar] [Green Version]
- Nakabeppu, Y; Kajitani, K; Sakamoto, K; Yamaguchi, H; Tsuchimoto, D. MTH1, an oxidized purine nucleoside triphosphatase, prevents the cytotoxicity and neurotoxicity of oxidized purine nucleotides. DNA Repair (Amst.) 2006, 5, 761–772. [Google Scholar] [Green Version]
- Ito, R; Hayakawa, H; Sekiguchi, M; Ishibashi, T. Multiple enzyme activities of Escherichia coli MutT protein for sanitization of DNA and RNA precursor pools. Biochemistry (Mosc). 2005, 44, 6670–6674. [Google Scholar] [Green Version]
- Furuta, A; Iida, T; Nakabeppu, Y; Iwaki, T. Expression of hMTH1 in the hippocampi of control and Alzheimer's disease. Neuroreport 2001, 12, 2895–2899. [Google Scholar] [Green Version]
- Shimura-Miura, H; Hattori, N; Kang, D; Miyako, K; Nakabeppu, Y; Mizuno, Y. Increased 8-oxo-dGTPase in the mitochondria of substantia nigral neurons in Parkinson's disease. Ann. Neurol. 1999, 46, 920–924. [Google Scholar] [Green Version]
- Hayakawa, H; Kuwano, M; Sekiguchi, M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry (Mosc). 2001, 40, 9977–9982. [Google Scholar] [Green Version]
- Hayakawa, H; Sekiguchi, M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry (Mosc). 2006, 45, 6749–6755. [Google Scholar] [Green Version]
- Hayakawa, H; Uchiumi, T; Fukuda, T; Ashizuka, M; Kohno, K; Kuwano, M; Sekiguchi, M. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry (Mosc). 2002, 41, 12739–12744. [Google Scholar] [Green Version]
- Yang, WH; Bloch, DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA 2007, 13, 704–712. [Google Scholar] [Green Version]
Share and Cite
Castellani, R.J.; Nunomura, A.; Rolston, R.K.; Moreira, P.I.; Takeda, A.; Perry, G.; Smith, M.A. Sublethal RNA Oxidation as a Mechanism for Neurodegenerative Disease. Int. J. Mol. Sci. 2008, 9, 789-806. https://doi.org/10.3390/ijms9050789
Castellani RJ, Nunomura A, Rolston RK, Moreira PI, Takeda A, Perry G, Smith MA. Sublethal RNA Oxidation as a Mechanism for Neurodegenerative Disease. International Journal of Molecular Sciences. 2008; 9(5):789-806. https://doi.org/10.3390/ijms9050789
Chicago/Turabian StyleCastellani, Rudy J., Akihiko Nunomura, Raj K. Rolston, Paula I. Moreira, Atsushi Takeda, George Perry, and Mark A. Smith. 2008. "Sublethal RNA Oxidation as a Mechanism for Neurodegenerative Disease" International Journal of Molecular Sciences 9, no. 5: 789-806. https://doi.org/10.3390/ijms9050789
APA StyleCastellani, R. J., Nunomura, A., Rolston, R. K., Moreira, P. I., Takeda, A., Perry, G., & Smith, M. A. (2008). Sublethal RNA Oxidation as a Mechanism for Neurodegenerative Disease. International Journal of Molecular Sciences, 9(5), 789-806. https://doi.org/10.3390/ijms9050789






