Development of a Generic PCR Detection of 3-Acetyldeoxynivalenol-, 15-Acetyldeoxynivalenol- and Nivalenol-Chemotypes of Fusarium graminearum Clade
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fusarium Strains
2.2. DNA Extraction
2.3. Primer Design
2.4. PCR Amplification
2.5. GC/MS Analysis
3. Results and Discussion
4. Conclusions
Acknowledgments
References
- Windels, CE. Economic and social impacts of Fusarium head blight: Changing farms and rural communities in the Northern Great Plains. Phytopathology 2000, 90, 17–21. [Google Scholar]
- Placinta, EM; D’Mello, JPF; Machdonald, AMC. A review of worldwide contamination of cereal grains and animal feed with Fusarium mycotoxins. Anim. Feed Sci. Tech 1999, 78, 21–37. [Google Scholar]
- Cleveland, TE; Dowd, PF; Desjardins, AE; Bhatnagar, D; Cotty, PJ. Unitied States department of agriculture-agricultural research service research on pre-harvest prevention of mycotoxins and mycotoxingenic fungi in US crops. Pest Manag. Sci 2003, 59, 629–642. [Google Scholar]
- Desjardins, AE; Manandhar, G; Plattner, RD; Maragos, CM; Shrestha, K; McCormick, SP. Occurrence of Fusarium species and mycotoxins in Nepalese maize and wheat and effect of traditional processing methods on mycotoxin levels. J. Agri. Food Chem 2000, 48, 1377–1383. [Google Scholar]
- D’Mello, JPF; Placinta, CM; Macdonald, AMC. Fusarium mycotoxins: A review of global implications for animal health, welfare and productivity. Anim. Feed Sci.Technol 1999, 80, 183–205. [Google Scholar]
- Gutleb, AC; Morrison, E; Murk, AJ. Cytotoxicity assays for mycotoxins produced by Fusarium strains: A review. Environ. Toxicol. Pharmacol 2002, 11, 309–320. [Google Scholar]
- Ward, TJ; Bielawski, JP; Kistler, HC; Sullivan, E; O’Donnell, K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 2002, 9, 9278–9283. [Google Scholar]
- Miller, JD; Greenhalgh, R; Wang, YZ; Lu, M. Trichothecene chemotypes of three Fusarium species. Mycologia 1991, 83, 121–130. [Google Scholar]
- Moss, MO; Thrane, U. Fusarium taxonomy with relation to trichothecene formation. Toxicol. Lett 2004, 153, 23–28. [Google Scholar]
- Haratian, M; Sharifnabi, B; Alizadeh, A; Safaie, N. PCR analysis of the Tri13 gene to determine the genetic potential of Fusarium graminearum isolates from Iran to produce Nivalenol and Deoxynivalenol. Mycopathologia 2008, 166, 109–116. [Google Scholar]
- O’Donnell, K; Kistler, HC; Tacke, BK; Casper, HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar]
- Grove, JF. The trichothecenes and their biosynthesis. In Fortschritte der Chemie organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products; Herz, W, Falk, H, Kirby, GW, Eds.; Springer: Wien, New York, USA, 2007; Volume 88, pp. 63–130. [Google Scholar]
- Ward, TJ; Bielawski, JP; Kistler, HC; Sullivan, E; O’Donnell, K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 2002, 9, 9278–9283. [Google Scholar]
- Nicholson, P; Simpson, DR; Wilson, AH; Chandler, E; Thomsett, M. Detection and differentiation of trichothecene and enniatin-producing Fusarium species on small-grain cereals. Eur. J. Plant Pathol 2004, 110, 503–514. [Google Scholar]
- Hsueh, CC; Liu, Y; Freund, MS. Indirect electrochemical detection of type-B trichothecene mycotoxins. Anal. Chem 1999, 71, 4075–4080. [Google Scholar]
- Schollenberger, M; Drochner, W. Fusarium toxins of the scirpentriol subgroup: A review. Mycopathologia 2007, 164, 101–118. [Google Scholar]
- Lee, T; Han, YK; Kim, KH; Yun, SH; Lee, YW. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol 2002, 5, 2148–2154. [Google Scholar]
- Hohn, TM; McCormick, SP; Desjardins, AE. Evidence for a gene cluster involving trichothecene-pathway biosynthetic genes in Fusarium sporotrichioides. Curr. Genet 1993, 24, 291–295. [Google Scholar]
- Brown, DW; McCormick, SP; Alexander, NJ; Proctor, RH; Desjardins, AE. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol 2001, 32, 121–133. [Google Scholar]
- Ji, L; Cao, K; Hu, T; Wang, S. Determination of deoxynivalenol and nivalenol chemotypes of Fusarium graminearum isolates from China by PCR assay. Phytopathology 2007, 155, 505–512. [Google Scholar]
- Qu, B; Li, HP; Zhang, JB; Xu, YB; Huang, T; Wu, AB; Zhao, CS; Carter, J; Nicholson, P; Liao, YC. Geographic distribution and genetic diversity of Fusarium graminearum and F. asiaticum on wheat spikes throughout China. Plant Pathol 2008, 57, 15–24. [Google Scholar]
- Carter, JP; Rezanoor, HN; Holden, D; Desjardins, AE; Plattner, RD; Nicholson, P. Variation in pathogenicity associated with the genetic diversity of Fusarium graminearum. Eur. J. Plant Pathol 2002, 108, 573–583. [Google Scholar]
- Nicholson, P; Rezanoor, HN; Simpson, DR; Joyce, D. Differentiation and quantification of the cereal eyespot fungi Tapesia yallundae and Tapesia acuformis using a PCR assay. Plant Pathol 1997, 46, 842–856. [Google Scholar]
- Li, HP; Wu, AB; Zhao, CS; Scholten, O; Löffler, H; Liao, YC. Development of a generic PCR detection of deoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum. FEMS Microbiol. Lett 2005, 243, 505–511. [Google Scholar]
- Busko, M; Ckhełowski, J; Popiel, D; Perkowski, J. Solid substrate bioassay to evaluate impact of Trichoderma on trichothecene mycotoxin production by Fusarium species. J. Sci. Food Agric 2008, 88, 536–541. [Google Scholar]
- Zhang, JB; Li, HP; Dang, FJ; Qu, B; Xu, YB; Zhao, CS; Liao, YC. Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol. Res 2007, 111, 967–975. [Google Scholar]
- Qu, B; Li, HP; Zhang, JB; Huang, T; Carter, J; Liao, YC; Nicholson, P. Comparison of genetic diversity and pathogenicity of Fusarium head blight pathogens from China and Europe by SSCP and seedling assays on wheat. Plant Pathol 2008, 57, 642–651. [Google Scholar]
- Chandler, EA; Simpson, DR; Thomsett, MA; Nicholson, P. Development of PCR assays to Tri7 and Tri13 trichothecene biosynthetic genes, and characterisation of chemotypes of Fusarium graminearum, Fusarium culmorum and Fusarium cerealis. Physiol. Mol. Plant Pathol 2003, 62, 355–367. [Google Scholar]
- Kimura, M; Takeshi, T; Naoko, TA; Shuichi, O; Makoto, F. Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci. Biotechnol. Biochem 2007, 71, 2105–2123. [Google Scholar]
- Brown, DW; McCormick, SP; Alexander, NJ; Proctor, RH; Desjardins, AE. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol 2002, 36, 224–233. [Google Scholar]
- Kimura, M; Tokai, T; O’Donnell, K; Ward, TJ; Fujimura, M; Hamamoto, H; Shibata, T; Yamaguchi, I. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett 2003, 539, 105–110. [Google Scholar]
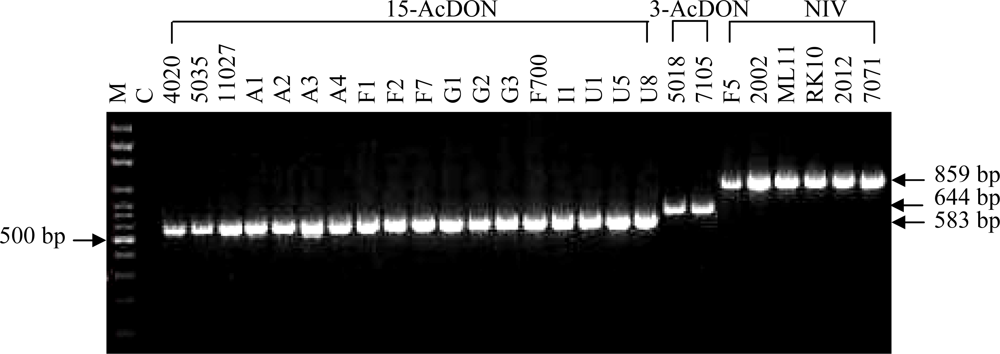
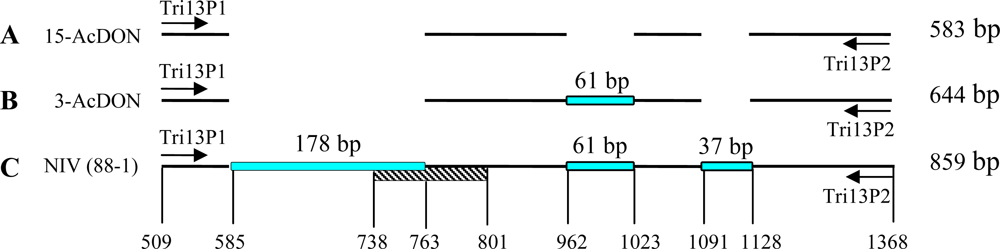
| Strain code | Origin | Host | Chemotype | PCR assay results
| |||
|---|---|---|---|---|---|---|---|
| Fragment size (bp) | NIV | 3-Ac DON | 15-Ac DON | ||||
| 2002 | China | Wheat | NIV a | 859 | + | − | − |
| 2012 | China | Wheat | NIV a | 859 | + | − | − |
| 3002 | China | Wheat | 15-AcDON a | 583 | − | − | + |
| 4020 | China | Wheat | 15-AcDON b | 583 | − | − | + |
| 5018 | China | Wheat | 3-AcDON c | 644 | − | + | − |
| 5035 | China | Wheat | 15-AcDON d | 583 | − | − | + |
| 5039 | China | Wheat | 3-AcDON a | 644 | − | + | − |
| 5119 | China | Wheat | 15-AcDON a | 583 | − | − | + |
| 5226 | China | Wheat | n.t. | 583 | − | − | + |
| 7105 | China | Wheat | 3-AcDON d | 644 | − | + | − |
| 7047 | China | Wheat | 3-AcDON a | 644 | − | + | − |
| 7071 | China | Wheat | NIV a | 859 | + | − | − |
| 7089 | China | Wheat | 3-AcDON a | 644 | − | + | − |
| 11027 | China | Wheat | 15-AcDON a | 583 | − | − | + |
| 12002 | China | Wheat | NIV a | 859 | + | − | − |
| 12003 | China | Wheat | NIV a | 859 | + | − | − |
| 13081 | China | Wheat | n.t. | 644 | − | + | − |
| 104 | China | Wheat | n.t. | 583 | − | − | + |
| CH1-1 | China | Wheat | n.t. | 583 | − | − | + |
| CH2-1 | China | Wheat | n.t. | 583 | − | − | + |
| SH | China | Maize | n.t. | 583 | − | − | + |
| LY-11 | China | Wheat | n.t. | 583 | − | − | + |
| YZ-2 | China | Rice | n.t. | 583 | − | − | + |
| JYH | China | Maize | n.t. | 583 | − | − | + |
| F1 | France | Wheat | 15-AcDON c | 583 | − | − | + |
| F2 | France | Wheat | 15-AcDON c | 583 | − | − | + |
| F4 | France | Wheat | 15-AcDON c | 583 | − | − | + |
| F5 | France | Wheat | n.t. | 859 | + | − | − |
| F6 | France | Wheat | NIV c | 859 | + | − | − |
| F7 | France | Wheat | 15-AcDON c | 583 | − | − | + |
| D5 | Germany | Wheat | DON e | 583 | − | − | + |
| G1 | Germany | Wheat | 15-AcDON c | 583 | − | − | + |
| G2 | Germany | Wheat | 15-AcDON c | 583 | − | − | + |
| G3 | Germany | Wheat | 15-AcDON c | 583 | − | − | + |
| F700 | Germany | Wheat | 15-AcDON b | 583 | − | − | + |
| G6 | Germany | Wheat | 15-AcDON c | 583 | − | − | + |
| I1 | Italy | Wheat | 15-AcDON c | 583 | − | − | + |
| I3 | Italy | Wheat | 15-AcDON c | 583 | − | − | + |
| ML11 | Nepal | Maize | NIV e | 859 | + | − | − |
| RK10(HKM215) | Nepal | Rice | NIV e | 859 | + | − | − |
| N6 (MK6) | Nepal | Maize | NIV e | 859 | + | − | − |
| U1 | UK | Wheat | 15-AcDON c | 583 | − | − | + |
| U2 | UK | Wheat | n.t. | 859 | + | − | − |
| U4 | UK | Wheat | 15-AcDON c | 583 | − | − | + |
| U5 | UK | Wheat | 15-AcDON c | 583 | − | − | + |
| U7 | UK | Wheat | n.t. | 583 | − | − | + |
| U8 | UK | Wheat | 15-AcDON c | 583 | − | − | + |
| UK1 | UK | Wheat | DON e | 583 | − | − | + |
| A1 | USA | Wheat | 15-AcDON c | 583 | − | − | + |
| A2 | USA | Wheat | 15-AcDON c | 583 | − | − | + |
| A3 | USA | Wheat | 15-AcDON c | 583 | − | − | + |
| A4 | USA | Wheat | 15-AcDON c | 583 | − | − | + |
| A5 (IL42) | USA | Wheat | 15-AcDON c | 583 | − | − | + |
© 2008 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/). This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, J.-H.; Li, H.-P.; Qu, B.; Zhang, J.-B.; Huang, T.; Chen, F.-F.; Liao, Y.-C. Development of a Generic PCR Detection of 3-Acetyldeoxynivalenol-, 15-Acetyldeoxynivalenol- and Nivalenol-Chemotypes of Fusarium graminearum Clade. Int. J. Mol. Sci. 2008, 9, 2495-2504. https://doi.org/10.3390/ijms9122495
Wang J-H, Li H-P, Qu B, Zhang J-B, Huang T, Chen F-F, Liao Y-C. Development of a Generic PCR Detection of 3-Acetyldeoxynivalenol-, 15-Acetyldeoxynivalenol- and Nivalenol-Chemotypes of Fusarium graminearum Clade. International Journal of Molecular Sciences. 2008; 9(12):2495-2504. https://doi.org/10.3390/ijms9122495
Chicago/Turabian StyleWang, Jian-Hua, He-Ping Li, Bo Qu, Jing-Bo Zhang, Tao Huang, Fang-Fang Chen, and Yu-Cai Liao. 2008. "Development of a Generic PCR Detection of 3-Acetyldeoxynivalenol-, 15-Acetyldeoxynivalenol- and Nivalenol-Chemotypes of Fusarium graminearum Clade" International Journal of Molecular Sciences 9, no. 12: 2495-2504. https://doi.org/10.3390/ijms9122495




