Evaluation of the Antimicrobial Activity of Cationic Peptides Loaded in Surface-Modified Nanoliposomes against Foodborne Bacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Peptides Design and Sequence Characteristics
2.2. Molecular Dynamic Simulation
2.3. Characterisation Physicochemical of Peptides in Aqueous Media
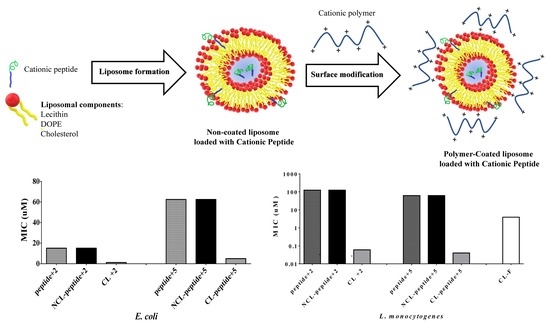
2.4. Polymer-Coated Liposome Coated with Peptides
2.5. Stability of Liposomes
2.6. Antimicrobial Activity
3. Materials and Methods
3.1. Bacterial Strains and Chemicals
3.2. Peptide Design
3.3. Peptide Synthesis
3.4. Molecular Dynamic Simulation
3.5. Physicochemical Characterisation of the Peptide in Aqueous Media
3.5.1. Surface Tension Measurements
3.5.2. Aggregation Index Measurements
3.6. Polymer-Coated Liposome Coated with Peptides
3.6.1. Preparation of Liposomes Loaded with Peptide
3.6.2. Liposome Surface Modification
3.6.3. Physicochemical Characterisation of Liposomes
3.6.4. Stability of Liposomes
3.7. Antimicrobial Activity
3.8. Statistical Analysis
4. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMP | Antimicrobial Peptide |
| FDA | Food and Drug Administration |
| MIC | Minimal Inhibition Concentration |
| Lys | Lysine |
| Ser | Serine |
| Arg | Arginine |
| Ala | Alanine |
| MD | Molecular Dynamic |
| RMSD | Root Mean Square Deviation |
| PDI | Polydispersity Index |
| NCL | non-coated liposomes |
| CL | coated liposomes |
| LPS | lipopolysaccharide |
| TBTU | 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium tetrafluoro-borate |
| DIEA | N,N-Diisopropylethylamine |
| DMF | Dimethylformamide |
| TFA | Trifluoroacetic acid |
| EDT | 1,2-Ethanedithiol |
| TIS | Triisopropylsilane |
| MHB | Mueller Hinton Broth |
| DOPE | dioleoyl-phosphatidyl-ethanolamine |
| PBCs | Periodic Boundary Conditions |
| PME | Particle Mesh Ewald |
References
- WHO. Estimates of the Global Burden of Foodborne Diseases; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Niaz, T.; Shabbir, S.; Noor, T.; Abbasi, R.; Raza, Z.A.; Imran, M. Polyelectrolyte multicomponent colloidosomes loaded with nisin Z for enhanced antimicrobial activity against foodborne resistant pathogens. Front. Microbiol. 2018, 8, 2700. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid. Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Garzón, J.; Ausili, A.; Manrique-Moreno, M.; Torrecillas, A.; Aranda, F.J.; Patiño, E.; Gomez-Fernández, J.C. The increase in positively charged residues in cecropin D-like Galleria mellonella favors its interaction with membrane models that imitate bacterial membranes. Arch. Biochem. Biophys. 2017, 629, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Hall, K.N.; Aguilar, M.I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic alpha helical antimicrobial peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Andreu, D.; Nogués, V.M.; Boix, E. Connecting peptide physicochemical and antimicrobial properties by a rational prediction model. PLoS ONE 2011, 6, e16968. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Demandt, A.; Nielsen, P.F.; Leprince, J.; Vaudry, H.; Woodhams, D.C. The alyteserins: Two families of antimicrobial peptides from the skin secretions of the midwife toad Alytes obstetricans (Alytidae). Peptides 2009, 30, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Ahmed, E.; Pal, T.; Sonnevend, A. Potent and rapid bactericidal action of alyteserin-1c and its [E4K] analog against multidrug-resistant strains of Acinetobacter baumannii. Peptides 2010, 31, 1806–1810. [Google Scholar] [CrossRef] [PubMed]
- Mojsoska, B.; Jenssen, H. Peptides and Peptidomimetics for Antimicrobial Drug Design. Pharmaceuticals 2015, 8, 366–415. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, M.; Giménez, B.; Da Silva, I.M.; Boelter, J.F.; Montero, P.; Gómez-Guillén, M.C.; Brandelli, A. Nanoencapsulation of an active peptidic fraction from sea bream scales collagen. Food Chem. 2014, 156, 144–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ron-Doitch, S.; Sawodny, B.; Kühbacher, A.; David, M.M.N.; Samanta, A.; Phopase, J.; Burger-Kentischer, A.; Griffith, M.; Golomb, G.; Rupp, S. Reduced cytotoxicity and enhanced bioactivity of cationic antimicrobial peptides liposomes in cell cultures and 3D epidermis model against HSV. J. Control. Release 2016, 229, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Alipour, M.; Halwani, M.; Omri, A.; Suntres, Z.E. Antimicrobial effectiveness of liposomal polymyxin B against resistant Gram-negative bacterial strains. Int. J. Pharm. 2008, 355, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, W.; Ye, A.; Peng, S.; Wei, F.; Liu, C.; Han, J. Environmental stress stability of microencapsules based on liposomes decorated with chitosan and sodium alginate. Food Chem. 2016, 196, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Kadian, S.S.; Harikumar, S.L. Eudragit and its Pharmaceutical Significance. Eudragit Pharm. Signif. 2009. [Google Scholar]
- Lopes, N.A.; Pinilla, C.M.B.; Brandelli, A. Pectin and polygalacturonic acid-coated liposomes as novel delivery system for nisin: Preparation, characterization and release behavior. Food Hydrocoll. 2017, 70, 1–7. [Google Scholar] [CrossRef]
- Gomaa, A.I.; Martinent, C.; Hammami, R.; Fliss, I.; Subirade, M. Dual Coating of Liposomes as Encapsulating Matrix of Antimicrobial Peptides: Development and Characterization. Front. Chem. 2017, 5, 103. [Google Scholar] [CrossRef] [PubMed]
- Pu, C.; Tang, W. A chitosan-coated liposome encapsulating antibacterial peptide, Apep10: Characterisation, triggered-release effects and antilisterial activity in thaw water of frozen chicken. Food Funct. 2016, 7, 4310–4322. [Google Scholar] [CrossRef]
- Da Silva, I.M.; Boelter, J.F.; Da Silveira, N.P.; Brandelli, A. Phosphatidylcholine nanovesicles coated with chitosan or chondroitin sulfate as novel devices for bacteriocin delivery. J. Nanopart. Res. 2014, 16, 2479. [Google Scholar] [CrossRef]
- Bordo, D.; Argos, P. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 1991, 217, 721–729. [Google Scholar] [CrossRef] [Green Version]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins †. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Alías, M.; Ayuso-Tejedor, S.; Fernández-Recio, J.; Cativiela, C.; Sancho, J. Helix propensities of conformationally restricted amino acids. Non-natural substitutes for helix breaking proline and helix forming alanine. Org. Biomol. Chem. 2010, 8, 788–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G. Determination of solution structure and lipid micelle location of an engineered membrane peptide by using one NMR experiment and one sample. Biochim. Biophys. Acta 2007, 1768, 3271–3281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, D.J.; Kim, D.T.; Steinman, L.; Fathman, C.G.; Rothbard, J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000, 56, 318–325. [Google Scholar] [CrossRef]
- Lee, D.L.; Mant, C.T.; Hodges, R.S. A novel method to measure self-association of small amphipathic molecules: Temperature profiling in reversed-phase chromatography. J. Biol. Chem. 2003, 278, 22918–22927. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, S.S.; Wang, S.; Dachineni, R.; Averineni, R.K.; Yang, Y.; Yin, H.; Bhat, G.J.; Guan, X. In Vitro and In Vivo Tumor Growth Inhibition by Glutathione Disulfide Liposomes. Cancer Growth Metastasis 2017. [Google Scholar] [CrossRef]
- Goldenbogen, B.; Brodersen, N.; Gramatica, A.; Loew, M.; Liebscher, J.; Herrmann, A.; Egger, H.; Budde, B.; Arbuzova, A. Reduction-sensitive liposomes from a multifunctional lipid conjugate and natural phospholipids: Reduction and release kinetics and cellular uptake. Langmuir 2011, 27, 10820–10829. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, S.S.; Xie, J.; Zhang, H.; Perumal, O.; Guan, X. Glutathione disulfide liposomes—A research tool for the study of glutathione disulfide associated functions and dysfunctions. Biochem. Biophys. Rep. 2016, 7, 225–229. [Google Scholar] [CrossRef]
- Hancock, R.E. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Gregoriadis, G. Liposome technology, Vol II Entrapment of drugs and other materials into liposomes [Internet]. Third edit. Vol. I. Informa Healthcare USA Inc. 1984. Available online: http://www.crcnetbase.com.ezlibproxy1.ntu.edu.sg/isbn/978-0-8493-8828-6 (accessed on 20 January 2019).
- Daraee, H.; Etemadi, A.; Kouhi, M.; Alimirzalu, S.; Akbarzadeh, A. Application of liposomes in medicine and drug delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 381–391. [Google Scholar] [CrossRef]
- Oñate-Garzón, J.; Manrique-Moreno, M.; Trier, S.; Leidy, C.; Torres, R.; Patiño, E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. 2017, 70, 238–245. [Google Scholar] [PubMed]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers 2008, 90, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Abraham, T.; Lewis, R.N.; Hodges, R.S.; McElhaney, R.N. Isothermal titration calorimetry studies of the binding of a rationally designed analogue of the antimicrobial peptide gramicidin s to phospholipid bilayer membranes. Biochemistry 2005, 44, 2103–2112. [Google Scholar] [CrossRef] [PubMed]
- Sonnenfeld, E.M.; Beveridge, T.J.; Koch, A.L.; Doyle, R.J. Asymmetric distribution of charge on the cell wall of Bacillus subtilis. J. Bacteriol. 1985, 163, 1167–1171. [Google Scholar] [PubMed]
- Alasino, R.V.; Ausar, S.F.; Bianco, I.D.; Castagna, L.F.; Contigiani, M.; Beltramo, D.M. Amphipathic and membrane-destabilizing properties of the cationic acrylate polymer Eudragit® E100. Macromol. Biosci. 2005, 5, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Romero, V.L.; Pons, P.; Bocco, J.L.; Manzo, R.H.; Alovero, F.L. Eudragit E100?? potentiates the bactericidal action of ofloxacin against fluoroquinolone-resistant Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2012, 334, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Briuglia, M.L.; Rotella, C.; McFarlane, A.; Lamprou, D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, D.A.; Nagle, J.F. Metastability in the phase behavior of dimyristoylphosphatidylethanolamine bilayers. Biochemistry 1984, 23, 1538–1541. [Google Scholar] [CrossRef]
- Fauchere, J.; Pliska, V. Hydrophobic parameters {pi} of amino-acid side chains from the partitioning of N-acetyl-amino-acid amides. Eur. J. Med. Chem. 1983, 8, 369–375. [Google Scholar]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci USA 1984, 81, 140–144. [Google Scholar] [CrossRef]
- Batcho, P.F.; Case, D.A.; Schlick, T. Optimized particle-mesh Ewald/multiple-time step integration for molecular dynamics simulations. J. Chem. Phys. 2001, 115, 4003–4018. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, J.D.; Neeson, M.J.; Dagastine, R.R.; Chan, D.Y.C.; Tabor, R.F. Measurement of surface and interfacial tension using pendant drop tensiometry. J. Colloid Interface Sci. 2015, 454, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Kahl, H.; Wadewitz, T.; Winkelmann, J. Surface tension of pure liquids and binary liquid mixtures. J. Chem. Eng. Data 2003, 48, 580–586. [Google Scholar] [CrossRef]
- Arévalo, M.L.; Yarce, J.C.; Oñate-Garzón, J.; Salamanca, H.C. Decrease of Antimicrobial Resistance through Polyelectrolyte-Coated Nanoliposomes Loaded with β-Lactam Drug. Pharmaceuticals 2018, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- CLSI, M07-A10: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition., CLSI (Clinical Lab. Stand. Institute). (2015). [CrossRef]
- Adetunji, V.O.; Adegoke, G.O. Formation of biofilm by strains of Listeria monocytogenes isolated from soft cheese ‘wara’ and its processing environment. J. Biotechnol. 2008, 7, 2893–2897. [Google Scholar]






| Name | Sequence | Q | <H> | <µH> | MW |
|---|---|---|---|---|---|
| 1 10 20 | |||||
| peptide +2 | H2N- GLKEIFKAGLGSLVKGIAAHVAS–COOH | +2 | 0.461 | 0.38 | 2266.7 |
| peptide +5 | H2N- GLKRIFKSGLGKLVKGISAHVAS–COOH | +5 | 0.373 | 0.434 | 2366.9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantor, S.; Vargas, L.; Rojas A., O.E.; Yarce, C.J.; Salamanca, C.H.; Oñate-Garzón, J. Evaluation of the Antimicrobial Activity of Cationic Peptides Loaded in Surface-Modified Nanoliposomes against Foodborne Bacteria. Int. J. Mol. Sci. 2019, 20, 680. https://doi.org/10.3390/ijms20030680
Cantor S, Vargas L, Rojas A. OE, Yarce CJ, Salamanca CH, Oñate-Garzón J. Evaluation of the Antimicrobial Activity of Cationic Peptides Loaded in Surface-Modified Nanoliposomes against Foodborne Bacteria. International Journal of Molecular Sciences. 2019; 20(3):680. https://doi.org/10.3390/ijms20030680
Chicago/Turabian StyleCantor, Stefania, Lina Vargas, Oscar E. Rojas A., Cristhian J. Yarce, Constain H. Salamanca, and Jose Oñate-Garzón. 2019. "Evaluation of the Antimicrobial Activity of Cationic Peptides Loaded in Surface-Modified Nanoliposomes against Foodborne Bacteria" International Journal of Molecular Sciences 20, no. 3: 680. https://doi.org/10.3390/ijms20030680