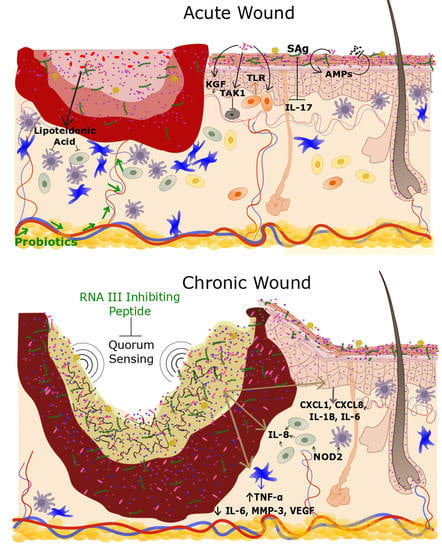
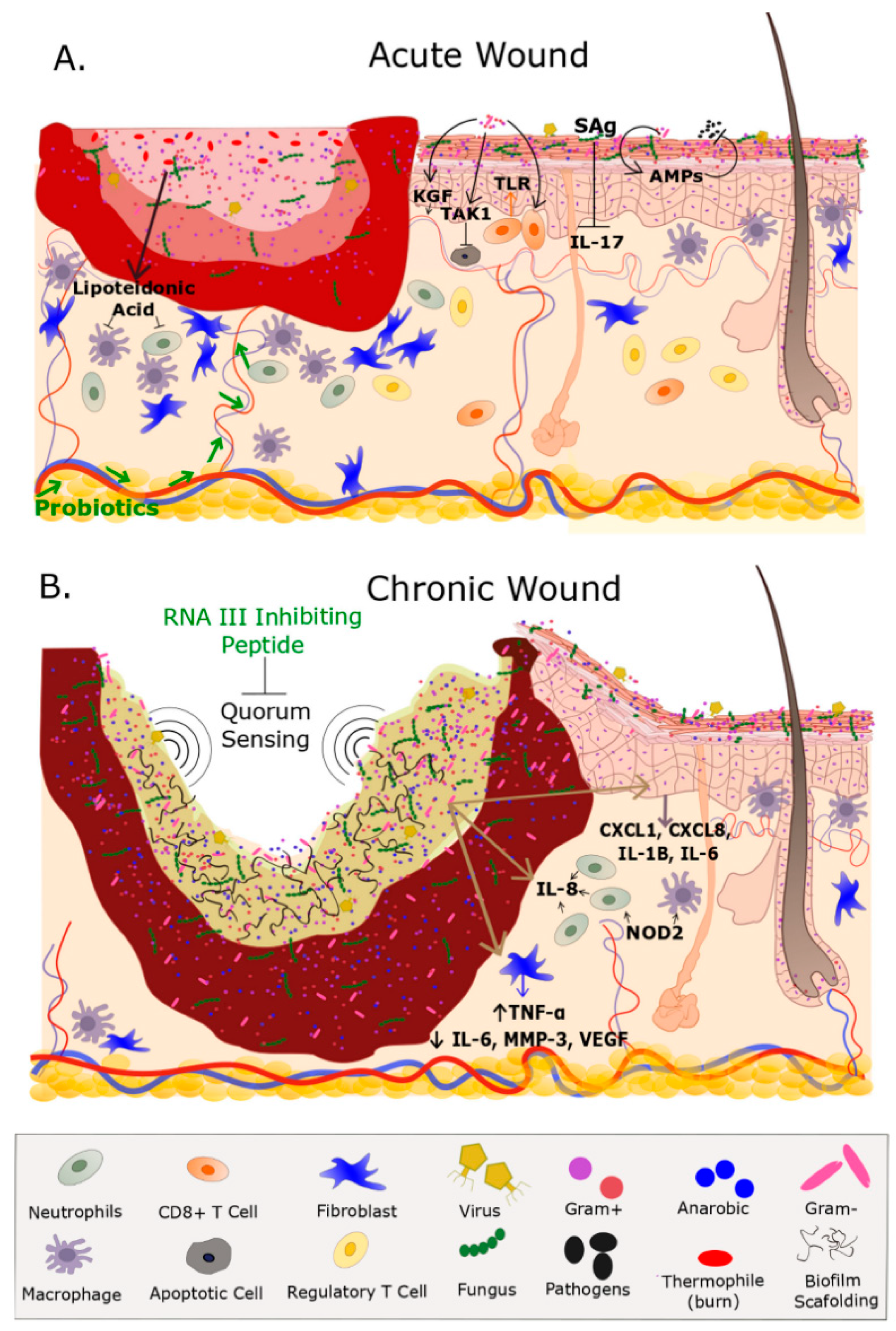
The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing
Abstract
1. Introduction
2. Defining the Healthy Skin Microbiome
2.1. Bacteria
2.2. Fungi
2.3. Viruses
2.4. Other Factors Defining the Skin Microbiome
3. The Cutaneous Microbiome: Aberrations in Human Wounds
3.1. Chronic Wounds
3.2. Acute Wounds
4. The Microbiome, Wound Healing and Inflammation: Mechanistic Insight from Model Systems
Effects of Bacterial Colonization on Skin Inflammation and Cutaneous Homeostasis
5. Modulating the Microbiome: Clinical Implications for Wound Healing and Tissue Regeneration
6. Conclusions
Funding
Conflicts of Interest
Abbreviations
| AKT/PKB | Protein Kinase B |
| AMP | Anti-Microbial Peptide |
| CXCL | Chemokine Ligand |
| DFU | Diabetic Foot Ulcer |
| DNA | Deoxyribonucleic Acid |
| DU | Decubitus Ulcer |
| EGF ® | Epidermal Growth Factor (Receptor) |
| ERK | Extracellular Signal Regulated Kinase |
| FOXP3 | Forkhead Box P3 Protein |
| GAS | Group A Streptococcus |
| hBD | Human Beta Defense Protein |
| HPV | Human Papilloma Virus |
| IL | Interleukin |
| KGF | Keratinocyte Growth Factor |
| MAP | Mitogen Activated Pathway |
| MAPK | Mitogen Activated Pathway Kinase |
| MKK | Mitogen Activated Pathway Kinase Kinase |
| MMP | Matrix Metalloproteinase |
| MRSA | Methicillin Resistant Staphylococcus aureus |
| NF-κβ | Nuclear Factor Kappa Beta |
| NOD | Nucleotide-binding oligomerization domain-containing protein |
| PI3K | phosphatidylinositol 3-kinase |
| rDNA | Ribosomal Deoxyribonucleic Acid |
| RegIIIy | Regenerating Islet Derived Protein Gamma |
| RNA | Ribonucleic Acid |
| Rnase | Ribonuclease |
| ROS | Reactive Oxygen Species |
| Sag | Superantigen |
| SCFA | Short Chain Fatty Acid |
| TAK | Transforming Growth Factor (TGF) F β-activated kinase |
| TGF | Transforming Growth Factor |
| TLR | Toll Like Receptor |
| TNF | Tumor Necrosis Factor |
| TRAF | Tumor Necrosis Factor (TNF) receptor-associated factor |
| TRAP | Target of RNAIII activating protein |
| Treg | T regulatory Cell |
| VEGF | Vascular Endothelial Growth Factor |
| VLU | Venous Leg Ulcer |
References
- DuPont, A.W.; DuPont, H.L. The intestinal microbiota and chronic disorders of the gut. Nature reviews. Gastroenterol. Hepatol. 2011, 8, 523–531. [Google Scholar]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. The commensal microbiota and the development of human disease—An introduction. J. Oral Microbiol. 2015, 7, 29128. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.R.; Gallo, R.L. The role of the skin microbiome in atopic dermatitis. Curr. Allergy Asthma Rep. 2015, 15, 65. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A. The skin microbiome: Potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin. Cutan. Med. Surg. 2014, 33, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Canesso, M.C.; Vieira, A.T.; Castro, T.B.; Schirmer, B.G.; Cisalpino, D.; Martins, F.S.; Rachid, M.A.; Nicoli, J.R.; Teixeira, M.M.; Barcelos, L.S. Skin wound healing is accelerated and scarless in the absence of commensal microbiota. J. Immunol. 2014, 193, 5171–5180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jiang, Z.; Li, D.; Jiang, D.; Wu, Y.; Ren, H.; Peng, H.; Lai, Y. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microbi. Ecol. 2015, 69, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- SanMiguel, A.; Grice, E.A. Interactions between host factors and the skin microbiome. Cell. Mol. Life Sci. 2015, 72, 1499–1515. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.A.; Smith, W.M.; Johnston, E.A.; Giblett, E.R. Bacterial flora of the normal human skin. J. Investig. Dermatol. 1950, 15, 305–324. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nature reviews. Microbiology 2011, 9, 244–253. [Google Scholar] [PubMed]
- Nakatsuji, T.; Chiang, H.I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 2013, 4, 1431. [Google Scholar] [CrossRef] [PubMed]
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Kong, H.H.; et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013, 498, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Paulino, L.C.; Tseng, C.H.; Blaser, M.J. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 2008, 8, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Paulino, L.C.; Tseng, C.H.; Strober, B.E.; Blaser, M.J. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 2006, 44, 2933–2941. [Google Scholar] [CrossRef] [PubMed]
- Cottam, E.M.; Wadsworth, J.; Knowles, N.J.; King, D.P. Full sequencing of viral genomes: Practical strategies used for the amplification and characterization of foot-and-mouth disease virus. Methods Mol. Biol. 2009, 551, 217–230. [Google Scholar] [PubMed]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 2015, 6, e01578. [Google Scholar] [CrossRef] [PubMed]
- Houldcroft, C.J.; Beale, M.A.; Breuer, J. Clinical and biological insights from viral genome sequencing. Nature reviews. Microbiology 2017, 15, 183–192. [Google Scholar] [PubMed]
- Krishnamurthy, S.R.; Wang, D. Origins and challenges of viral dark matter. Virus Res. 2017, 239, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Antonsson, A.; Erfurt, C.; Hazard, K.; Holmgren, V.; Simon, M.; Kataoka, A.; Hossain, S.; Hakangard, C.; Hansson, B.G. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J. Gen. Virol. 2003, 84, 1881–1886. [Google Scholar] [CrossRef] [PubMed]
- Antonsson, A.; Forslund, O.; Ekberg, H.; Sterner, G.; Hansson, B.G. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 2000, 74, 11636–11641. [Google Scholar] [CrossRef] [PubMed]
- Astori, G.; Lavergne, D.; Benton, C.; Hockmayr, B.; Egawa, K.; Garbe, C.; de Villiers, E.M. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J. Investig. Dermatol. 1998, 110, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Foulongne, V.; Sauvage, V.; Hebert, C.; Dereure, O.; Cheval, J.; Gouilh, M.A.; Pariente, K.; Segondy, M.; Burguiere, A.; Manuguerra, J.C.; et al. Human skin microbiota: High diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE 2012, 7, e38499. [Google Scholar] [CrossRef] [PubMed]
- Jonczyk-Matysiak, E.; Weber-Dabrowska, B.; Zaczek, M.; Miedzybrodzki, R.; Letkiewicz, S.; Lusiak-Szelchowska, M.; Gorski, A. Prospects of Phage Application in the Treatment of Acne Caused by Propionibacterium acnes. Front. Microbiol. 2017, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Landini, M.M.; Borgogna, C.; Peretti, A.; Doorbar, J.; Griffin, H.; Mignone, F.; Lai, A.; Urbinati, L.; Matteelli, A.; Gariglio, M.; et al. Identification of the skin virome in a boy with widespread human papillomavirus-2-positive warts that completely regressed after administration of tetravalent human papillomavirus vaccine. Br. J. Dermatol. 2015, 173, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.E.; Tsao, H. The skin microbiome: Current perspectives and future challenges. J. Am. Acad. Dermatol. 2013, 69, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Estrada, I.; Gao, Z.; Clemente, J.C.; Costello, E.K.; Knight, R. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013, 7, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zeng, D.N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.X. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PLoS ONE 2015, 10, e0141842. [Google Scholar] [CrossRef] [PubMed]
- Hospodsky, D.; Pickering, A.J.; Julian, T.R.; Miller, D.; Gorthala, S.; Boehm, A.B.; Peccia, J. Hand bacterial communities vary across two different human populations. Microbiology 2014, 160, 1144–1152. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.H.; Wilkins, D.; Lee, P.K. Insights into the pan-microbiome: Skin microbial communities of Chinese individuals differ from other racial groups. Sci. Rep. 2015, 5, 11845. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.J.; Clark, R.A. Cutaneous wound healing. N. Engl. J. Med. 1999, 341, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Thorne, C.; Chung, K.C.; Gosain, A.; Guntner, G.C.; Mehrara, B.J. Grabb and Smith’s Plastic Surgery; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2014. [Google Scholar]
- Zeeuwen, P.L.; Kleerebezem, M.; Timmerman, H.M.; Schalkwijk, J. Microbiome and skin diseases. Curr. Opin. Allergy Clin. Immunol. 2013, 13, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, T.; Beasley, D.E.; Hedenec, P.; Xiao, Z.; Zhang, S.; Li, J.; Lin, Q.; Li, X. Diet Diversity Is Associated with Beta but not Alpha Diversity of Pika Gut Microbiota. Front. Microbiol. 2016, 7, 1169. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Walker, A.W.; Capilla, A.; Benitez-Paez, A.; Palau, F.; Parkhill, J.; Castillejo, G.; Sanz, Y. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome 2018, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Seite, S.; Flores, G.E.; Henley, J.B.; Martin, R.; Zelenkova, H.; Aguilar, L.; Fierer, N. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J. Drugs Dermatol. 2014, 13, 1365–1372. [Google Scholar] [PubMed]
- Brandwein, M.; Steinberg, D.; Meshner, S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes 2016, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K.; Gordillo, G.M.; Roy, S.; Kirsner, R.; Lambert, L.; Hunt, T.K.; Gottrup, F.; Gurtner, G.C.; Longaker, M.T. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair Regen. 2009, 17, 763–771. [Google Scholar] [CrossRef] [PubMed]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Thomson, C.H. Biofilms: Do they affect wound healing? Int. Wound J. 2011, 8, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.K.; Wolcott, R.; Wolk, D.M.; Bharara, M.; Kimbriel, H.R.; Armstrong, D.G. Diabetic foot infections: A need for innovative assessments. Int. J. Lower Extremity Wounds 2010, 9, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Ammons, M.C.; Morrissey, K.; Tripet, B.P.; Van Leuven, J.T.; Han, A.; Lazarus, G.S.; Zenilman, J.M.; Stewart, P.S.; James, G.A.; Copie, V. Biochemical association of metabolic profile and microbiome in chronic pressure ulcer wounds. PLoS ONE 2015, 10, e0126735. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, M.; Vicaretti, M.; Sparks, J.; Bansal, S.; Bush, S.; Liu, M.; Darling, A.; Harry, E.; Burke, C.M. A longitudinal study of the diabetic skin and wound microbiome. PeerJ 2017, 5, e3543. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Antolin, M.; Santos, J.; Torrejon, A.; Casellas, F.; Borruel, N.; Guarner, F.; Malagelada, J.R. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am. J. Gastroenterol. 2008, 103, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Loesche, M.; Gardner, S.E.; Kalan, L.; Horwinski, J.; Zheng, Q.; Hodkinson, B.P.; Tyldsley, A.S.; Franciscus, C.L.; Hillis, S.L.; Mehta, S.; et al. Temporal Stability in Chronic Wound Microbiota Is Associated with Poor Healing. J. Investig. Dermatol. 2017, 137, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.; Loesche, M.; Hodkinson, B.P.; Heilmann, K.; Ruthel, G.; Gardner, S.E.; Grice, E.A. Redefining the Chronic-Wound Microbiome: Fungal Communities Are Prevalent, Dynamic, and Associated with Delayed Healing. mBio 2016, 7, e01058-16. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.; Grice, E.A. Fungi in the Wound Microbiome. Adv. Wound Care 2018, 7, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Plichta, J.K.; Gao, X.; Lin, H.; Dong, Q.; Toh, E.; Nelson, D.E.; Gamelli, R.L.; Grice, E.A.; Radek, K.A. Cutaneous Burn Injury Promotes Shifts in the Bacterial Microbiome in Autologous Donor Skin: Implications for Skin Grafting Outcomes. Shock 2017, 48, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Huang, Y.C.; Chen, L.Y.; Yu, S.C.; Yu, H.Y.; Chuang, S.S. The skin microbiome of wound scars and unaffected skin in patients with moderate to severe burns in the subacute phase. Wound Repair Regen. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bartow-McKenney, C.; Hannigan, G.D.; Horwinski, J.; Hesketh, P.; Horan, A.D.; Mehta, S.; Grice, E.A. The microbiota of traumatic, open fracture wounds is associated with mechanism of injury. Wound Repair Regen. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hannigan, G.D.; Hodkinson, B.P.; McGinnis, K.; Tyldsley, A.S.; Anari, J.B.; Horan, A.D.; Grice, E.A.; Mehta, S. Culture-independent pilot study of microbiota colonizing open fractures and association with severity, mechanism, location, and complication from presentation to early outpatient follow-up. J. Orthop. Res. 2014, 32, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Kirker, K.R.; Secor, P.R.; James, G.A.; Fleckman, P.; Olerud, J.E.; Stewart, P.S. Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair Regen. 2009, 17, 690–699. [Google Scholar] [CrossRef] [PubMed]
- De Breij, A.; Haisma, E.M.; Rietveld, M.; El Ghalbzouri, A.; van den Broek, P.J.; Dijkshoorn, L.; Nibbering, P.H. Three-dimensional human skin equivalent as a tool to study Acinetobacter baumannii colonization. Antimicrob. Agents Chemother. 2012, 56, 2459–2464. [Google Scholar] [CrossRef] [PubMed]
- Den Reijer, P.M.; Haisma, E.M.; Lemmens-den Toom, N.A.; Willemse, J.; Koning, R.I.; Demmers, J.A.; Dekkers, D.H.; Rijkers, E.; El Ghalbzouri, A.; Nibbering, P.H.; et al. Detection of Alpha-Toxin and Other Virulence Factors in Biofilms of Staphylococcus aureus on Polystyrene and a Human Epidermal Model. PLoS ONE 2016, 11, e0145722. [Google Scholar]
- Clark, R.A. Wound repair. Curr. Opin. Cell Biol. 1989, 1, 1000–1008. [Google Scholar] [CrossRef]
- Kirker, K.R.; James, G.A.; Fleckman, P.; Olerud, J.E.; Stewart, P.S. Differential effects of planktonic and biofilm MRSA on human fibroblasts. Wound Repair Regen. 2012, 20, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef] [PubMed]
- Takao, K.; Miyakawa, T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2015, 112, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Sorkin, M.; Glotzbach, J.P.; Longaker, M.T.; Gurtner, G.C. Surgical approaches to create murine models of human wound healing. J. Biomed. Biotechnol. 2011, 2011, 969618. [Google Scholar] [CrossRef] [PubMed]
- Seaton, M.; Hocking, A.; Gibran, N.S. Porcine models of cutaneous wound healing. ILAR J. 2015, 56, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, T.P.; Eaglstein, W.H.; Davis, S.C.; Mertz, P. The pig as a model for human wound healing. Wound Repair Regen. 2001, 9, 66–76. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, M.K.; Peacock, T.J.; Akers, K.S.; Burmeister, D.M. Initial Characterization of the Pig Skin Bacteriome and Its Effect on In Vitro Models of Wound Healing. PLoS ONE 2016, 11, e0166176. [Google Scholar] [CrossRef] [PubMed]
- Chiarello, M.; Villeger, S.; Bouvier, C.; Auguet, J.C.; Bouvier, T. Captive bottlenose dolphins and killer whales harbor a species-specific skin microbiota that varies among individuals. Sci. Rep. 2017, 7, 15269. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Glatz, M.; Horiuchi, K.; Kawasaki, H.; Akiyama, H.; Kaplan, D.H.; Kong, H.H.; Amagai, M.; Nagao, K. Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 2015, 42, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Schierle, C.F.; De la Garza, M.; Mustoe, T.A.; Galiano, R.D. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 2009, 17, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef] [PubMed]
- Plichta, J.K.; Droho, S.; Curtis, B.J.; Patel, P.; Gamelli, R.L.; Radek, K.A. Local burn injury impairs epithelial permeability and antimicrobial peptide barrier function in distal unburned skin. Crit. Care Med. 2014, 42, e420–e431. [Google Scholar] [CrossRef] [PubMed]
- Plichta, J.K.; Holmes, C.J.; Gamelli, R.L.; Radek, K.A. Local Burn Injury Promotes Defects in the Epidermal Lipid and Antimicrobial Peptide Barriers in Human Autograft Skin and Burn Margin: Implications for Burn Wound Healing and Graft Survival. J. Burn Care Res. 2017, 38, e212–e226. [Google Scholar] [CrossRef] [PubMed]
- Linehan, J.L.; Harrison, O.J.; Han, S.J.; Byrd, A.L.; Vujkovic-Cvijin, I.; Villarino, A.V.; Sen, S.K.; Shaik, J.; Smelkinson, M.; Tamoutounour, S.; et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 2018, 172, 784–796.e18. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Bouladoux, N.; Linehan, J.L.; Han, S.J.; Harrison, O.J.; Wilhelm, C.; Conlan, S.; Himmelfarb, S.; Byrd, A.L.; Deming, C.; et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015, 520, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Di Nardo, A.; Nakatsuji, T.; Leichtle, A.; Yang, Y.; Cogen, A.L.; Wu, Z.R.; Hooper, L.V.; Schmidt, R.R.; von Aulock, S.; et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 2009, 15, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Midorikawa, K.; Ouhara, K.; Komatsuzawa, H.; Kawai, T.; Yamada, S.; Fujiwara, T.; Yamazaki, K.; Sayama, K.; Taubman, M.A.; Kurihara, H.; et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 2003, 71, 3730–3739. [Google Scholar] [CrossRef] [PubMed]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Gotz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.O.; Dale, B.A. Innate immune response of oral and foreskin keratinocytes: Utilization of different signaling pathways by various bacterial species. Infect. Immun. 2004, 72, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K.; Karau, M.J.; Greenwood-Quaintance, K.E.; Tilahun, A.Y.; Krogman, A.; David, C.S.; Pritt, B.S.; Patel, R.; Rajagopalan, G. Superantigen-Producing Staphylococcus aureus Elicits Systemic Immune Activation in a Murine Wound Colonization Model. Toxins 2015, 7, 5308–5319. [Google Scholar] [CrossRef] [PubMed]
- Vojtov, N.; Ross, H.F.; Novick, R.P. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc. Natl. Acad. Sci. USA 2002, 99, 10102–10107. [Google Scholar] [CrossRef] [PubMed]
- Secor, P.R.; James, G.A.; Fleckman, P.; Olerud, J.E.; McInnerney, K.; Stewart, P.S. Staphylococcus aureus Biofilm and Planktonic cultures differentially impact gene expression, mapk phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol. 2011, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.P.; Merryman, M.S.; Harris-Arnold, A.; McKinney, S.A.; Seidel, C.W.; Loethen, S.; Proctor, K.N.; Guo, L.; Sanchez Alvarado, A. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. elife 2016, 5, e16793. [Google Scholar] [CrossRef] [PubMed]
- Wilgus, T.A. Immune cells in the healing skin wound: Influential players at each stage of repair. Pharmacol. Res. 2008, 58, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.; Campbell, L.; Crompton, R.A.; Singh, G.; McHugh, B.J.; Davidson, D.J.; McBain, A.J.; Cruickshank, S.M.; Hardman, M.J. Microbial Host Interactions and Impaired Wound Healing in Mice and Humans: Defining a Role for BD14 and NOD2. J. Investig. Dermatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Radek, K.A. Antimicrobial anxiety: The impact of stress on antimicrobial immunity. J. Leukoc. Biol. 2010, 88, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Clinton, A.; Carter, T. Chronic Wound Biofilms: Pathogenesis and Potential Therapies. Lab. Med. 2015, 46, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Scalise, A.; Bianchi, A.; Tartaglione, C.; Bolletta, E.; Pierangeli, M.; Torresetti, M.; Marazzi, M.; Di Benedetto, G. Microenvironment and microbiology of skin wounds: The role of bacterial biofilms and related factors. Semin. Vasc. Surg. 2015, 28, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Hochwalt, P.C.; Usui, M.L.; Underwood, R.A.; Singh, P.K.; James, G.A.; Stewart, P.S.; Fleckman, P.; Olerud, J.E. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: A model for the study of chronic wounds. Wound Repair Regen. 2010, 18, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Cirioni, O.; Giacometti, A.; Ghiselli, R.; Braunstein, J.B.; Silvestri, C.; Mocchegiani, F.; Saba, V.; Scalise, G. Treatment of Staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob. Agents Chemother. 2007, 51, 2226–2229. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Meneghin, A.; Hogaboam, C.M. Infectious disease, the innate immune response, and fibrosis. J. Clin. Investig. 2007, 117, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Starr, C.R.; Engleberg, N.C. Role of hyaluronidase in subcutaneous spread and growth of group A streptococcus. Infect. Immun. 2006, 74, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Blanchet-Rethore, S.; Bourdes, V.; Mercenier, A.; Haddar, C.H.; Verhoeven, P.O.; Andres, P. Effect of a lotion containing the heat-treated probiotic strain Lactobacillus johnsonii NCC 533 on Staphylococcus aureus colonization in atopic dermatitis. Clin. Cosmet. Investig. Dermatol. 2017, 10, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Two, A.M.; Chun, K.A.; Narala, S.; Geha, R.S.; Hata, T.R.; Gallo, R.L. Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression. J. Investig. Dermatol. 2016, 136, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Gueniche, A.; Knaudt, B.; Schuck, E.; Volz, T.; Bastien, P.; Martin, R.; Rocken, M.; Breton, L.; Biedermann, T. Effects of nonpathogenic gram-negative bacterium Vitreoscilla filiformis lysate on atopic dermatitis: A prospective, randomized, double-blind, placebo-controlled clinical study. Br. J. Dermatol. 2008, 159, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Valdez, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigon, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Satish, L.; Gallo, P.H.; Johnson, S.; Yates, C.C.; Kathju, S. Local Probiotic Therapy with Lactobacillus plantarum Mitigates Scar Formation in Rabbits after Burn Injury and Infection. Surg. Infect. 2017, 18, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, S.; Bayani, M.; Bahmani, F.; Tajabadi-Ebrahimi, M.; Bayani, M.A.; Jafari, P.; Asemi, Z. The beneficial effects of probiotic administration on wound healing and metabolic status in patients with diabetic foot ulcer: A randomized, double-blind, placebo-controlled trial. Diabetes/Metab. Res. Rev. 2018, 34, e2970. [Google Scholar] [CrossRef] [PubMed]
- Argenta, A.; Satish, L.; Gallo, P.; Liu, F.; Kathju, S. Local Application of Probiotic Bacteria Prophylaxes against Sepsis and Death Resulting from Burn Wound Infection. PLoS ONE 2016, 11, e0165294. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Butler, E.; Lindholm, C.; Nilson, B.; Michanek, P.; Vasquez, A. Fighting Off Wound Pathogens in Horses with Honeybee Lactic Acid Bacteria. Curr. Microbiol. 2016, 73, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Middelkoop, E.; van den Bogaerdt, A.J.; Lamme, E.N.; Hoekstra, M.J.; Brandsma, K.; Ulrich, M.M. Porcine wound models for skin substitution and burn treatment. Biomaterials 2004, 25, 1559–1567. [Google Scholar] [CrossRef]
- Kasatpibal, N.; Whitney, J.D.; Saokaew, S.; Kengkla, K.; Heitkemper, M.M.; Apisarnthanarak, A. Effectiveness of Probiotic, Prebiotic, and Synbiotic Therapies in Reducing Postoperative Complications: A Systematic Review and Network Meta-analysis. Clin. Infect. Dis. 2017, 64, 1531S–S1560. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Bruhs, A.; Schwarz, T. The Short-Chain Fatty Acid Sodium Butyrate Functions as a Regulator of the Skin Immune System. J. Investig. Dermatol. 2017, 137, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Kao, M.S.; Huang, S.; Chang, W.L.; Hsieh, M.F.; Huang, C.J.; Gallo, R.L.; Huang, C.M. Microbiome precision editing: Using PEG as a selective fermentation initiator against methicillin-resistant Staphylococcus aureus. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Poutahidis, T.; Kearney, S.M.; Levkovich, T.; Qi, P.; Varian, B.J.; Lakritz, J.R.; Ibrahim, Y.M.; Chatzigiagkos, A.; Alm, E.J.; Erdman, S.E. Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PLoS ONE 2013, 8, e78898. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.H.; van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325. [Google Scholar] [CrossRef] [PubMed]
- Scutera, S.; Zucca, M.; Savoia, D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin. Drug Discov. 2014, 9, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.A.; Bates, N.C.; Hancock, R.E. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 1986, 29, 496–500. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [PubMed]
- Monnappa, A.K.; Dwidar, M.; Seo, J.K.; Hur, J.H.; Mitchell, R.J. Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Sci. Rep. 2014, 4, 3811. [Google Scholar] [CrossRef] [PubMed]
| Bacteria | Positive Effects | Negative Effects | Associated Signaling Pathways |
|---|---|---|---|
| Staphylococcus epidermidis | Stimulates keratinocyte production of host AMPs (hBD3, RNase7) [22,74,75,88] Induces CD8+ T and IL-17A+ T cells [79] Enhances innate barrier immunity and limits pathogen invasion in absence of inflammation [6,74,80,89] | Occasionally pathogenic Implicated in production of biofilms [79,90,91,92,93] | NF-κB [74,76,89] TRAF1 [7] TLR2/CD36/CD14-p38, MAPK [7] EGFR TRAP [70] |
| Staphylococcus aureus | At a local level, super antigen production results in less skin inflammation and purulence due to decreased production of exotoxins and neutrophilic chemotactic factors [81] Amplifies innate immune response of skin via production of AMPs (hBD-3, hBD2, LL-37, RNAse7) [78,79] | Usually pathogenic Implicated in production of biofilms and delayed wound healing in chronic wounds [91,92] Super antigen production elicits robust activation of immune system [81] | TRAP [70,94] phosphatidylinositol 3-kinase/AKT NF/kB ERK TLR-2 [9] |
| Group A streptococcus (GAS) | Stimulates production of AMPs, promote epithelial differentiation [95] Activates plasminogen which promotes Keratinocyte chemotaxis and potential re-epithelization of wounds [95] | Usually pathogenic Express proteases which prevent neutrophil recruitment [79,96,97] Produces hyaluronidase which allows bacteria migration through host Extracellular matrix [7] Common cause of superficial and deep skin infections i.e., impetigo, erysipelas, cellulitis [6] | NF-κB/p65 [80] |
| Pseudomonas aeruginosa | Accelerates epithelialization and neovascularization in acute wounds Suppresses staphylococcal pathogens in polymicrobial wounds [84] | Usually pathogenic Implicated in production of biofilms and delayed wound healing in chronic wounds [90,91,92,93] | Nod2 [87] TAK1/MKK/p38 [85] |
| Corynebacterium jeikeium | Manganese acquisition and production of superoxide dismutase result in host epidermal protection from free radical oxygen species (ROS) [5] | Occasionally pathogenic Common cause of nosocomial skin infections [95] | N/A |
| Propionibacteria | Production of bacteriocins protect sebaceous ducts from other pathogenic inhabitants [77] induces expression of TLR2 and TLR4 in keratinocytes71 | Occasionally pathogenic Overabundance associated with development of Acne [5] | N/A |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnson, T.R.; Gómez, B.I.; McIntyre, M.K.; Dubick, M.A.; Christy, R.J.; Nicholson, S.E.; Burmeister, D.M. The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. Int. J. Mol. Sci. 2018, 19, 2699. https://doi.org/10.3390/ijms19092699
Johnson TR, Gómez BI, McIntyre MK, Dubick MA, Christy RJ, Nicholson SE, Burmeister DM. The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. International Journal of Molecular Sciences. 2018; 19(9):2699. https://doi.org/10.3390/ijms19092699
Chicago/Turabian StyleJohnson, Taylor R., Belinda I. Gómez, Matthew K. McIntyre, Michael A. Dubick, Robert J. Christy, Susannah E. Nicholson, and David M. Burmeister. 2018. "The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing" International Journal of Molecular Sciences 19, no. 9: 2699. https://doi.org/10.3390/ijms19092699
APA StyleJohnson, T. R., Gómez, B. I., McIntyre, M. K., Dubick, M. A., Christy, R. J., Nicholson, S. E., & Burmeister, D. M. (2018). The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. International Journal of Molecular Sciences, 19(9), 2699. https://doi.org/10.3390/ijms19092699