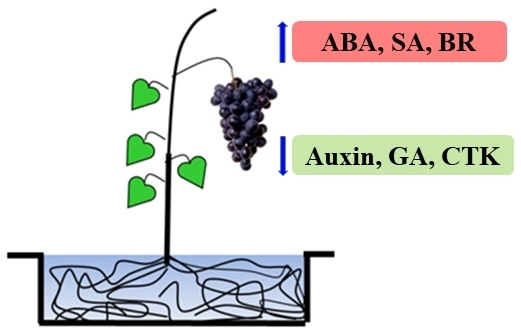
Transcriptomic Analyses of Root Restriction Effects on Phytohormone Content and Signal Transduction during Grape Berry Development and Ripening
Abstract
:1. Introduction
2. Results
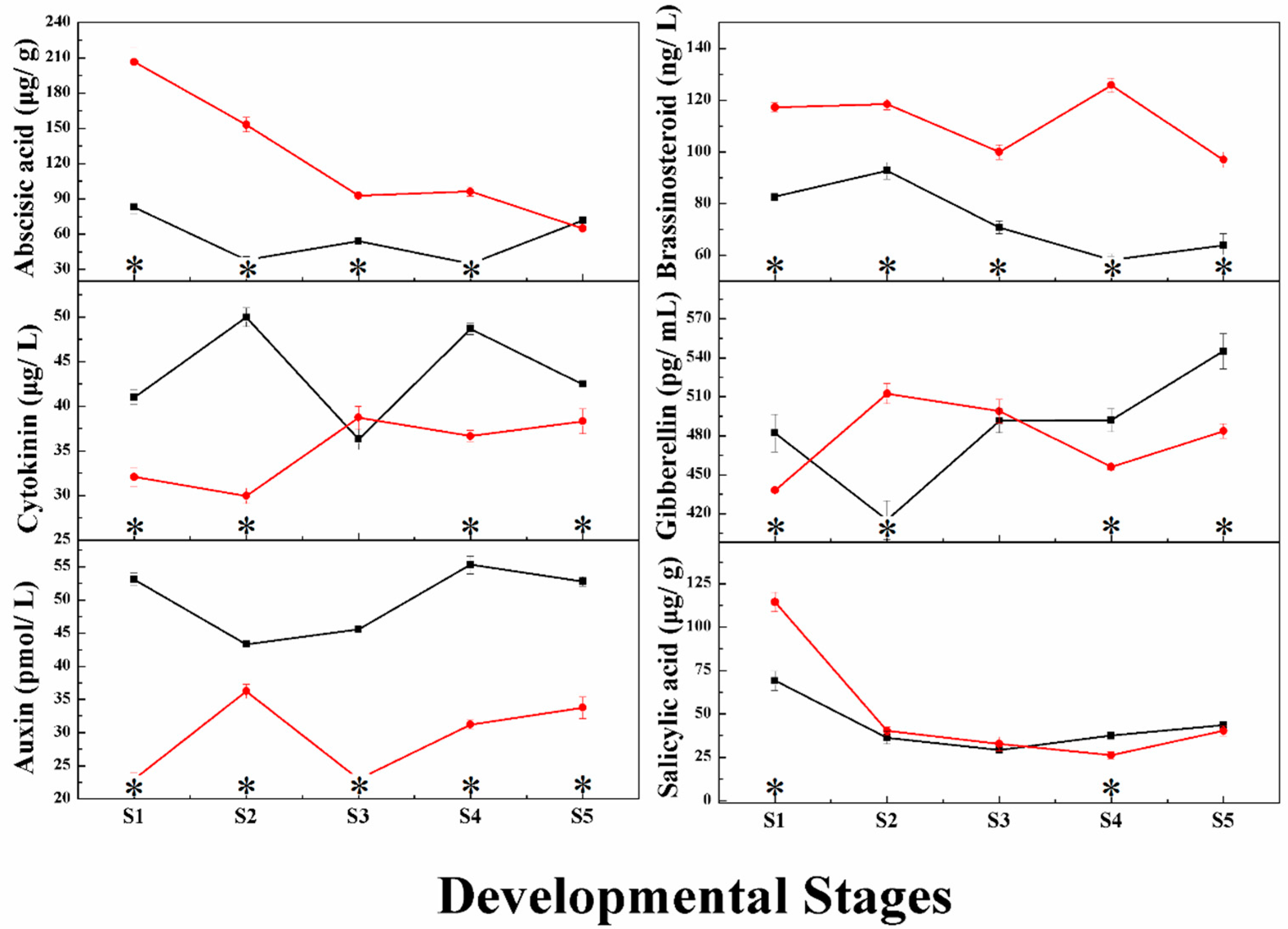
2.1. Changes in Phytohormone Content during Grape Berry Development
2.2. RNA-Sequencing Results
2.3. Phytohormone Biosynthesis and Signal Transduction Pathways
2.3.1. Auxin
2.3.2. Cytokinin
2.3.3. Gibberellin
2.3.4. Abscisic Acid
2.3.5. Ethylene
2.3.6. Brassinosteroid
2.3.7. Jasmonic Acid
2.3.8. Salicylic Acid
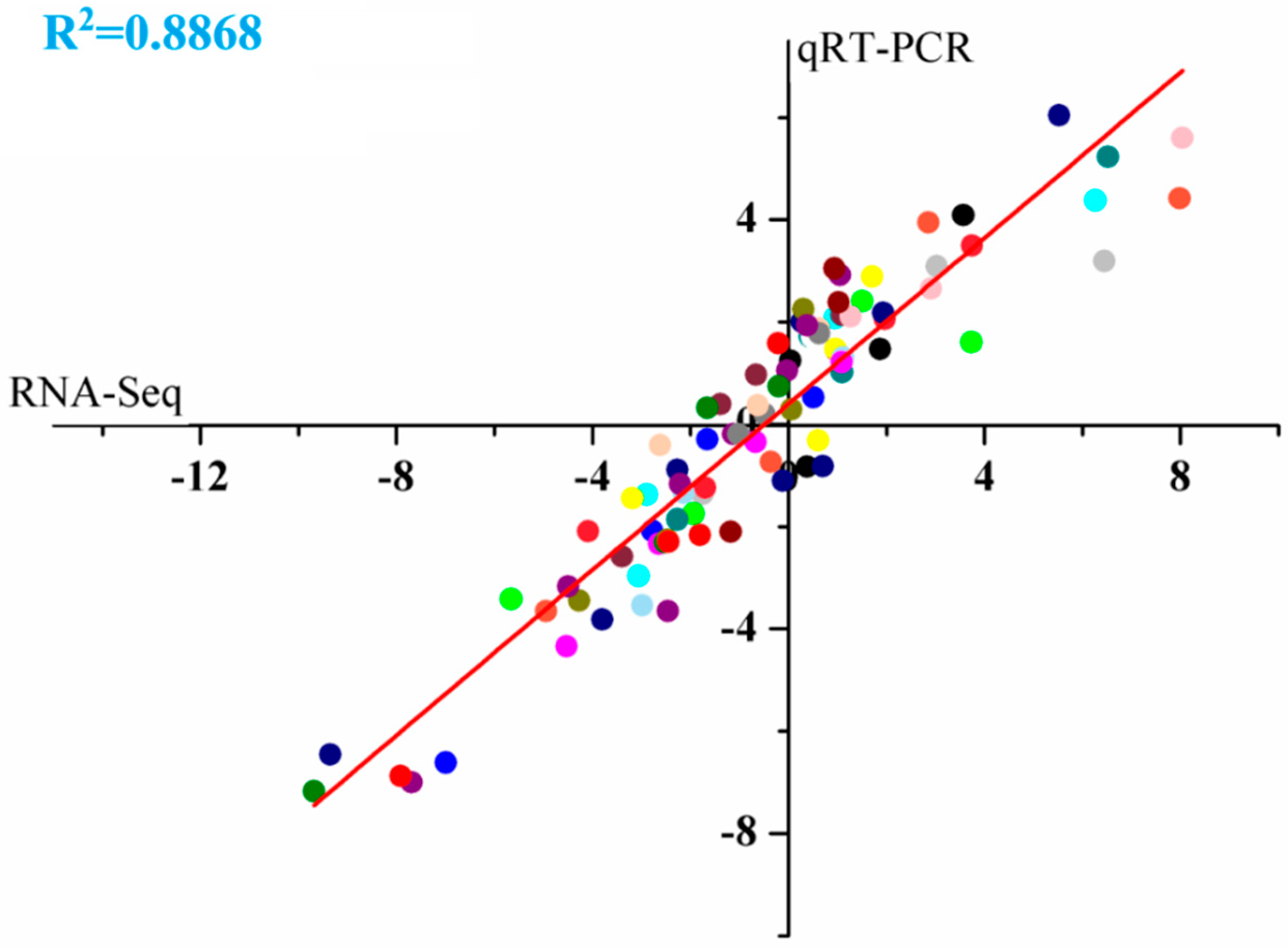
2.4. Validation of Different Gene Expression Using Real-Time Quantitative PCR (qRT-PCR)
3. Discussion
4. Materials and Methods
4.1. Materials Collected
4.2. Auxin, Cytokinin, Gibberellin and Brassinosteroid Quantification
4.3. Abscisic Acid and Salicylic Acid Quantification
4.4. RNA Extraction and RNA-Seq
4.5. Validation of Transcriptome Analysis Using qRT-PCR
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kuhn, N.; Delrot, S. Berry ripening: Recently heard through the grapevine. J. Exp. Bot. 2014, 65, 4543. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nat. Rev. 2009, 459, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhu, L.; Wang, S.; Gu, W.; Huang, D.; Xu, W.; Jiang, A.; Li, S. Nitrate uptake kinetics of grapevine under root restriction. Sci. Hortic. 2007, 111, 358–364. [Google Scholar] [CrossRef]
- Xie, Z.S.; Li, B.; Forney, C.F.; Xu, W.P.; Wang, S.P. Changes in sugar content and relative enzyme activity in grape berry in response to root restriction. Sci. Hortic. 2009, 123, 39–45. [Google Scholar] [CrossRef]
- Wang, S.; Okamoto, G.; Hirano, K.; Lu, J.; Zhang, C. Effects of restricted rooting volume on vine growth and berry development of kyoho grapevines. Am. J. Enol. Viticult. 2001, 52, 248–253. [Google Scholar]
- Wang, B.; He, J.; Yang, B.; Yu, X.; Li, J.; Zhang, C.; Xu, W.; Bai, X.; Cao, X.; Wang, S. Root restriction affected anthocyanin composition and up-regulated the transcription of their biosynthetic genes during berry development in ‘summer black’ grape. Acta Physiol. Plant. 2013, 35, 2205–2217. [Google Scholar] [CrossRef]
- Wang, B.; He, J.; Duan, C.; Yu, X.; Zhu, L.; Xie, Z.; Zhang, C.; Xu, W.; Wang, S. Root restriction affects anthocyanin accumulation and composition in berry skin of ‘kyoho’ grape (vitis vinifera L. × vitis labrusca L.) during ripening. Sci. Hortic. 2012, 137, 20–28. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed] [Green Version]
- Leng, F.; Lin, Q.; Wu, D.; Wang, S.; Wang, D.; Sun, C. Comparative Transcriptomic Analysis of Grape Berry in Response to Root Restriction during Developmental Stages. Molecules 2016, 21, 1431. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates scf(tir1)-dependent degradation of aux/iaa proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Jennifer, P.C.; Kieber, J.J. Cytokinin signaling: Two-components and more. Trends Plant Sci. 2008, 13, 85–92. [Google Scholar]
- Yamaguchi, S. Gibberellin biosynthesis in arabidopsis. Phytochem. Rev. 2006, 5, 39–47. [Google Scholar] [CrossRef]
- Hirano, K.; Ueguchitanaka, M.; Matsuoka, M. Gid1-mediated gibberellin signaling in plants. Trends Plant Sci. 2008, 13, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Gomi, K.; Sasaki, A.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Kitano, H.; Matsuoka, M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004, 37, 626–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, M.; Koshiba, T. Complex regulation of aba biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Klingler, J.P.; Batelli, G.; Zhu, J.K. Aba receptors: The start of a new paradigm in phytohormone signalling. J. Exp. Bot. 2010, 61, 3199–3210. [Google Scholar] [CrossRef] [PubMed]
- And, S.F.Y.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Biol. 1984, 35, 155–189. [Google Scholar]
- Ouaked, F.; Rozhon, W.; Lecourieux, D.; Hirt, H. A MAPK pathway mediates ethylene signaling in plants. EMBO J. 2003, 22, 1282–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsey, K.; Pullen, M.L.; Topping, J.F. Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci. 2003, 8, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Bancoş, S.; Nomura, T.; Sato, T.; Molnár, G.; Bishop, G.J.; Koncz, C.; Yokota, T.; Nagy, F.; Szekeres, M. Regulation of transcript levels of the arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002, 130, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, L.; Yin, Y. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathwaysf. Plant Physiol. 2011, 53, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mullet, J.E. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. 1997, 48, 355–381. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.; Gao, X.; Jones, A.D.; Howe, G.A. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in arabidopsis. Plant J. 2009, 59, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.I.; Leon, J.; Raskin, I. Biosynthesis and metabolism of salicylic acid. Proc. Natl. Acad. Sci. USA 1995, 92, 4076–4079. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Boden, E.; Arias, J. Salicylic acid and npr1 induce the recruitment of trans-activating tga factors to a defense gene promoter in arabidopsis. Plant Cell 2003, 15, 1846–1858. [Google Scholar] [CrossRef] [PubMed]
- Mcatee, P.; Karim, S.; Schaffer, R.; David, K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front. Plant Sci. 2013, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Symons, G.M.; Davies, C.; Shavrukov, Y.; Dry, I.B.; Reid, J.B.; Thomas, M.R. Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol. 2006, 140, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2014, 65, 4561–4575. [Google Scholar] [CrossRef] [PubMed]
- Fortes, A.M.; Teixeira, R.T.; Agudelo-Romero, P. Complex interplay of hormonal signals during grape berry ripening. Molecules 2015, 20, 9326. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchishinozaki, K.; Shinozaki, K.; Kakimoto, T. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, L.; Bing, Y.; Yangdong, G. The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 2014, 65, 4577. [Google Scholar]
- Davies, C.; Boss, P.K. Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 1997, 115, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Yamaguchishinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Deluc, L.G.; Quilici, D.R.; Decendit, A.; Grimplet, J.; Wheatley, M.D.; Schlauch, K.A.; Mérillon, J.M.; Cushman, J.C.; Cramer, G.R. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of cabernet sauvignon and chardonnay. BMC Genom. 2009, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Yamane, T.; Jeong, S.T.; Gotoyamamoto, N.; Koshita, Y.; Kobayashi, S. Effects of temperature on anthocyanin biosynthesis in grape berry skins. Am. J. Enol. Viticult. 2006, 57, 54–59. [Google Scholar]
- Toh, S.; Imamura, A.; Watanabe, A.; Nakabayashi, K.; Okamoto, M.; Jikumaru, Y.; Hanada, A.; Aso, Y.; Ishiyama, K.; Tamura, N. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in arabidopsis seeds. Plant Physiol. 2008, 146, 1368–1385. [Google Scholar] [CrossRef] [PubMed]
- Horvath, E.; Csiszar, J.; Galle, A.; Poor, P.; Szepesi, A.; Tari, I. Hardening with salicylic acid induces concentration-dependent changes in abscisic acid biosynthesis of tomato under salt stress. J. Plant. Physiol. 2015, 183, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Chervin, C.; Elkereamy, A.; Roustan, J.P.; Latche, A.; Lamon, J.; Bouzayen, M. Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci. 2004, 167, 1301–1305. [Google Scholar] [CrossRef] [Green Version]
- Deluc, L.G.; Jérôme, G.; Wheatley, M.D.; Tillett, R.L.; Quilici, D.R.; Craig, O.; Schooley, D.A.; Schlauch, K.A.; Cushman, J.C.; Cramer, G.R. Transcriptomic and metabolite analyses of cabernet sauvignon grape berry development. BMC Genom. 2007, 8, 429. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, G.A.; Matthews, M.A.; Shaghasi, T.H.; Mcelrone, A.J.; Castellarin, S.D. Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 2010, 232, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhang, C.; Pervaiz, T.; Zhao, P.; Liu, Z.; Wang, B.; Wang, C.; Zhang, L.; Fang, J.; Qian, J. Jasmonic acid involves in grape fruit ripening and resistant against botrytis cinerea. Funct. Integr. Genom. 2016, 16, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Niculcea, M.; Martinez-Lapuente, L.; Guadalupe, Z.; Sánchez-Díaz, M.; Morales, F.; Ayestarán, B.; Antolín, M.C. Effects of water-deficit irrigation on hormonal content and nitrogen compounds in developing berries of vitis vinifera L. cv. Tempranillo. J. Plant Growth Regul. 2013, 32, 551–563. [Google Scholar] [CrossRef]
- Wen, P.F.; Chen, J.Y.; Kong, W.F.; Pan, Q.H.; Wan, S.B.; Huang, W.D. Salicylic acid induced the expression of phenylalanine ammonia-lyase gene in grape berry. Plant Sci. 2005, 169, 928–934. [Google Scholar] [CrossRef]
- Liang, S.; Mei, Z.; Jie, R.; Qi, J.; Zhang, G.; Ping, L. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol. 2010, 10, 257. [Google Scholar]
- Shan, L.L.; Li, X.; Wang, P.; Cai, C.; Zhang, B.; Sun, C.D.; Zhang, W.S.; Xu, C.J.; Ferguson, I.; Chen, K.S. Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 2008, 227, 1243. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. Rseqc: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. Kobas 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Z.C.; Osada, T.; Glass, O.; Yang, X.Y.; Lei, G.; Lyerly, H.K.; Clay, T.M. Ligand-independent TLR signals generated by ectopic overexpression of myd88 generate local and systemic anti-tumor immunity. Cancer Res. 2010, 70, 7209. [Google Scholar] [CrossRef] [PubMed]


| Gene ID | Log2FC | Functional Annotation | ||||
|---|---|---|---|---|---|---|
| S1RR/S1Control | S2RR/S2Control | S3RR/S3Control | S4RR/S4Control | S5RR/S5Control | ||
| VIT_04s0044g01740 | −1.02 | hydroxymethylglutaryl-CoA reductase | ||||
| VIT_06s0004g01510 | 1.74 | lipoxygenase | ||||
| VIT_09s0002g01080 | 1.80 | −2.50 | lipoxygenase | |||
| VIT_02s0087g00910 | −1.88 | 9-cis-epoxycarotenoid dioxygenase | ||||
| VIT_10s0003g03750 | 1.82 | 9-cis-epoxycarotenoid dioxygenase | ||||
| VIT_19s0093g00550 | 3.72 | 9-cis-epoxycarotenoid dioxygenase | ||||
| VIT_14s0083g01110 | −2.15 | brassinosteroid-6-oxidase 2 | ||||
| VIT_19s0140g00120 | 1.15 | gibberellin 2-oxidase | ||||
| VIT_02s0025g00240 | 3.84 | 2.44 | beta-carotene 3-hydroxylase | |||
| VIT_16s0050g01090 | 1.59 | beta-carotene 3-hydroxylase | ||||
| VIT_03s0088g01150 | −1.22 | squalene monooxygenase | ||||
| VIT_11s0016g02380 | 3.15 | 2.55 | aminocyclopropanecarboxylate oxidase | |||
| VIT_12s0059g01380 | 1.66 | aminocyclopropanecarboxylate oxidase | ||||
| VIT_15s0048g00370 | −2.00 | −1.73 | transketolase | |||
| VIT_08s0007g05000 | 2.83 | S-adenosylmethionine synthetase | ||||
| VIT_12s0028g00960 | 1.99 | phytoene synthase | ||||
| VIT_00s0391g00070 | 1.30 | 3-deoxy-7-phosphoheptulonate synthase | ||||
| VIT_06s0004g02620 | −2.48 | 3.77 | 2.90 | phenylalanine ammonia-lyase | ||
| VIT_13s0019g04460 | −1.04 | 3.99 | 3.02 | phenylalanine ammonia-lyase | ||
| VIT_16s0039g01100 | −3.10 | phenylalanine ammonia-lyase | ||||
| VIT_16s0039g01110 | −2.64 | phenylalanine ammonia-lyase | ||||
| VIT_16s0039g01120 | −3.50 | phenylalanine ammonia-lyase | ||||
| VIT_09s0054g01090 | −1.11 | cycloartenol synthase | ||||
| VIT_01s0026g00940 | 1.96 | two-component response regulator ARR-A family | ||||
| VIT_08s0007g05390 | 1.71 | two-component response regulator ARR-A family | ||||
| VIT_13s0067g03070 | 4.18 | two-component response regulator ARR-A family | ||||
| VIT_18s0001g10450 | 1.27 | abscisic acid (ABA) responsive element binding factor | ||||
| VIT_05s0049g00510 | −3.69 | 1.84 | ethylene-responsive transcription factor 1 | |||
| VIT_07s0005g03230 | −1.66 | −4.62 | ethylene-responsive transcription factor 1 | |||
| VIT_07s0005g03260 | −2.23 | −3.89 | ethylene-responsive transcription factor 1 | |||
| VIT_06s0004g05240 | −2.43 | ethylene receptor | ||||
| VIT_05s0049g00090 | −1.92 | ethylene receptor | ||||
| VIT_07s0005g00090 | −2.77 | auxin-responsive GH3 gene family | ||||
| VIT_07s0104g00930 | 1.44 | gibberellin receptor GID1 | ||||
| VIT_05s0020g04670 | −1.81 | auxin-responsive protein IAA | ||||
| VIT_05s0020g04680 | −1.79 | auxin-responsive protein IAA | ||||
| VIT_05s0049g01970 | −1.61 | auxin-responsive protein IAA | ||||
| VIT_07s0141g00270 | 3.93 | auxin-responsive protein IAA | ||||
| VIT_09s0002g04080 | −2.65 | auxin-responsive protein IAA | ||||
| VIT_09s0002g05160 | 1.30 | auxin-responsive protein IAA | ||||
| VIT_11s0016g04490 | 2.32 | −1.52 | auxin-responsive protein IAA | |||
| VIT_14s0030g02310 | −1.11 | auxin-responsive protein IAA | ||||
| VIT_01s0146g00480 | 1.44 | jasmonate ZIM domain-containing protein | ||||
| VIT_09s0002g00890 | 1.21 | jasmonate ZIM domain-containing protein | ||||
| VIT_11s0016g00710 | 3.03 | jasmonate ZIM domain-containing protein | ||||
| VIT_06s0004g05460 | 2.14 | protein phosphatase 2C | ||||
| VIT_11s0016g03180 | 1.30 | protein phosphatase 2C | ||||
| VIT_13s0019g02200 | 1.67 | protein phosphatase 2C | ||||
| VIT_16s0050g02680 | 8.18 | 2.78 | 2.56 | protein phosphatase 2C | ||
| VIT_03s0088g00710 | 4.35 | 3.60 | pathogenesis-related protein 1 | |||
| VIT_03s0088g00810 | 2.32 | pathogenesis-related protein 1 | ||||
| VIT_02s0012g01270 | −1.26 | −2.19 | abscisic acid receptor PYR/PYL family | |||
| VIT_02s0154g00010 | 3.61 | −3.01 | SAUR family protein | |||
| VIT_04s0023g00580 | 2.69 | SAUR family protein | ||||
| VIT_15s0048g00530 | 1.51 | SAUR family protein | ||||
| VIT_16s0098g01150 | 1.70 | SAUR family protein | ||||
| VIT_11s0052g01190 | 1.89 | −4.88 | 2.07 | −3.95 | xyloglucosyl transferase TCH4 | |
| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| GAPDH | TGGAGCTGAATTTGTTGT | GTGGAGTTCTGGCTTGTA |
| VIT_06s0004g01510 | GACTGGCTTGGTAAAACACTCCT | GCACCAATCTCCCCAAAATC |
| VIT_06s0004g05460 | TCGCACTACTTCGGAGTTTATGA | ATCGCATTCTTCCAACCTGAC |
| VIT_10s0003g03750 | AACCAGATAACTCAGCCACCG | AAGAAGTGATGCCCAGCGAC |
| VIT_11s0016g02380 | CCCTGGAGGATAAAGAAACTGC | TTCTCCAAACCAAGATTCTCACA |
| VIT_13s0067g00330 | CTCTACACTTTCGGCGGACAC | CACAGCAGCATCACGGAATC |
| VIT_14s0083g01110 | AACATCCAGGAGAAAACCAAAGA | CCAGGAAGGTCAATAGGCAGA |
| VIT_15s0048g00370 | TTTAACCCCAAGAACCCCTACT | TGGCAATACCCTGACCAAGAG |
| VIT_16s0050g02680 | GAAATGCCAATCTGAGACCGA | CTCCTCCCAATCACCGACAC |
| VIT_18s0001g10450 | GGTGAGGGCAGGTGTAGTGAG | AAAATAGGTGATTGCGTGGAAA |
| VIT_19s0093g00550 | AACGCTGGGCTCGTCTATTT | CTCCACGTCTGGGGATTTGT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leng, F.; Cao, J.; Wang, S.; Jiang, L.; Li, X.; Sun, C. Transcriptomic Analyses of Root Restriction Effects on Phytohormone Content and Signal Transduction during Grape Berry Development and Ripening. Int. J. Mol. Sci. 2018, 19, 2300. https://doi.org/10.3390/ijms19082300
Leng F, Cao J, Wang S, Jiang L, Li X, Sun C. Transcriptomic Analyses of Root Restriction Effects on Phytohormone Content and Signal Transduction during Grape Berry Development and Ripening. International Journal of Molecular Sciences. 2018; 19(8):2300. https://doi.org/10.3390/ijms19082300
Chicago/Turabian StyleLeng, Feng, Jinping Cao, Shiping Wang, Ling Jiang, Xian Li, and Chongde Sun. 2018. "Transcriptomic Analyses of Root Restriction Effects on Phytohormone Content and Signal Transduction during Grape Berry Development and Ripening" International Journal of Molecular Sciences 19, no. 8: 2300. https://doi.org/10.3390/ijms19082300
APA StyleLeng, F., Cao, J., Wang, S., Jiang, L., Li, X., & Sun, C. (2018). Transcriptomic Analyses of Root Restriction Effects on Phytohormone Content and Signal Transduction during Grape Berry Development and Ripening. International Journal of Molecular Sciences, 19(8), 2300. https://doi.org/10.3390/ijms19082300