pSTAT3 Levels Have Divergent Expression Patterns and Associations with Survival in Squamous Cell Carcinoma and Adenocarcinoma of the Oesophagus
Abstract
:1. Introduction
2. Results
2.1. Divergent Associations of pSTAT3 and Survival in OAC versus SCC
2.2. IL-6R Expression in OAC and SCC
2.3. Functional Effects of STAT3 Inhibition on SCC and OAC In Vitro
2.3.1. Cell Viability
2.3.2. Cell Proliferation
2.3.3. Cell Migration
2.3.4. Apoptosis
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment
4.2. Tissue Microarray Construction
4.3. Immunohistochemistry
4.4. MTT Assay
4.5. BrdU Proliferation Assay
4.6. xCelligence Migration Assay
4.7. Annexin-V-FITC/Propidium Iodide Apoptosis Assay
4.8. Statistical Analysis
4.8.1. In Vitro
4.8.2. Ex Vivo
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chavan, S.; Bray, F.; Lortet-Tieulent, J.; Goodman, M.; Jemal, A. International variations in bladder cancer incidence and mortality. Eur. Urol. 2014, 66, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Torjesen, I. Large personalised medicine trial in lung cancer heralds new research partnership. BMJ 2014, 348. [Google Scholar] [CrossRef] [PubMed]
- Judd, L.M.; Menheniott, T.R.; Ling, H.; Jackson, C.B.; Howlett, M.; Kalantzis, A.; Priebe, W.; Giraud, A.S. Inhibition of the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and in vivo. PLoS ONE 2014, 9, e95993. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- White, C.A.; Nicola, N.A. SOCS3: An essential physiological inhibitor of signaling by interleukin-6 and G-CSF family cytokines. JAKSTAT 2013, 2, e25045. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef]
- Lapeire, L.; Hendrix, A.; Lambein, K.; Van Bockstal, M.; Braems, G.; Van Den Broecke, R.; Limame, R.; Mestdagh, P.; Vandesompele, J.; Vanhove, C.; et al. Cancer-associated adipose tissue promotes breast cancer progression by paracrine oncostatin M and Jak/STAT3 signaling. Cancer Res. 2014, 74, 6806–6819. [Google Scholar] [CrossRef] [PubMed]
- Hodge, D.R.; Hurt, E.M.; Farrar, W.L. The role of IL-6 and STAT3 in inflammation and cancer. Eur. J. Cancer 2005, 41, 2502–2512. [Google Scholar] [CrossRef] [PubMed]
- Pai, R.; Lin, C.; Tran, T.; Tarnawski, A. Leptin activates STAT and ERK2 pathways and induces gastric cancer cell proliferation. Biochem. Biophys. Res. Commun. 2005, 331, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.Q.; Ruan, X.; DU, X.; Shang, L.; Cai, Y.; Xu, X.; Wang, M.R.; Zhang, Y.; Fu, S.B. Overexpression of p-Stat3 and Mcl-1, and their correlation with differentiation and apoptotic resistance in esophageal squamous cell carcinoma. Zhonghua Zhong Liu Za Zhi 2013, 35, 579–584. [Google Scholar] [PubMed]
- You, Z.; Xu, D.; Ji, J.; Guo, W.; Zhu, W.; He, J. JAK/STAT signal pathway activation promotes progression and survival of human oesophageal squamous cell carcinoma. Clin. Transl. Oncol. 2012, 14, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Li, S.S.; Yan, A.H.; Lu, C.X.; Guo, Y.P. Constitutive activation of signal transducers and activators of transcription 3 and expression of its target gene products in human ESCC cell line. Nan Fang Yi Ke Da Xue Xue Bao 2006, 26, 441–444. (in Chinese). [Google Scholar] [PubMed]
- Renehan, A.G.; Tyson, M.; Egger, M.; Heller, R.F.; Zwahlen, M. Body-mass index and incidence of cancer: A systematic review and meta-analysis of prospective observational studies. Lancet 2008, 371, 569–578. [Google Scholar] [CrossRef]
- Timme, S.; Ihde, S.; Fichter, C.D.; Waehle, V.; Bogatyreva, L.; Atanasov, K.; Kohler, I.; Schöpflin, A.; Geddert, H.; Faller, G.; et al. STAT3 expression, activity and functional consequences of STAT3 inhibition in esophageal squamous cell carcinomas and Barrett’s adenocarcinomas. Oncogene 2014, 33, 3256–3266. [Google Scholar] [CrossRef] [PubMed]
- Schust, J.; Sperl, B.; Hollis, A.; Mayer, T.U.; Berg, T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006, 13, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Sellier, H.; Rébillard, A.; Guette, C.; Barré, B.; Coqueret, O. How should we define STAT3 as an oncogene and as a potential target for therapy? JAKSTAT, 2013, 2, e24716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, Y.A.; Jo, S.Y.; Lee, H.Y.; Lee, C. Inhibition of IL-6/STAT3 axis and targeting Axl and Tyro3 receptor tyrosine kinases by apigenin circumvent taxol resistance in ovarian cancer cells. Int. J. Oncol. 2015, 46, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Fofaria, N.M.; Srivastava, S.K. STAT3 induces anoikis resistance, promotes cell invasion and metastatic potential in pancreatic cancer cells. Carcinogenesis 2015, 36, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Levidou, G.; Sachanas, S.; Pangalis, G.; Kalpadakis, C.; Yiakoumis, X.; Moschogiannis, M.; Kyrtsonis, M.C.; Vassilakopoulos, T.; Tsirkinidis, P.; Kontopidou, F.; et al. Immunohistochemical analysis of IL-6, IL-8/CXCR2 axis, Tyr p-STAT-3, and SOCS-3 in lymph nodes from patients with chronic lymphocytic leukemia: Correlation between microvascular characteristics and prognostic significance. Biomed. Res. Int. 2014, 2014, 251479. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Huang, Z.; Zhao, J.; Han, Y.; Li, Z.; Zhang, Y.; Li, Y.; Chen, X.; Hu, X.; Zhao, H.; et al. Prognostic role of phosphor-STAT3 in patients with cancers of the digestive system: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0127356. [Google Scholar]
- Chen, X.; Ying, Z.; Lin, X.; Lin, H.; Wu, J.; Li, M.; Song, L. Acylglycerol kinase augments JAK2/STAT3 signaling in esphageal squamous cells. J. Clin. Investig. 2013, 123, 2576–2589. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xiao, W.; Ma, J.; Zhang, Y.; Li, R.; Ye, J.; Wang, X.; Zhong, X.; Wang, S. Dual high expression of STAT3 and cyclinD1 is associated with poor prognosis after curative resection of esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 7989–7998. [Google Scholar] [PubMed]
- Mesteri, I.; Schoppmann, S.F.; Preusser, M.; Birner, P. Overexpression of CMET is associated with signal transducer and activator of transcription 3 activation and diminished prognosis in oesophageal adenocarcinoma but not in squamous cell carcinoma. Eur. J. Cancer 2014, 50, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, C.; He, J.; Guo, Q.; Hu, D.; Yang, X.; Wang, J.; Kang, Y.; She, R.; Wang, Z.; et al. STAT3 inhibitor stattic enhances radiosensitivity in esophageal squamous cell carcinoma. Tumour Biol. 2014, 36, 2135–2142. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.R.; Wu, W.J.; Liu, S.X.; Zuo, L.F.; Wang, Y.; Yang, J.Z.; Nan, Y.M. Nimesulide inhibits the growth of human esophageal carcinoma cells by inactivating the JAK2/STAT3 pathway. Pathol. Res. Pract. 2015, 211, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Schoppmann, S.F.; Jesch, B.; Friedrich, J.; Jomrich, G.; Maroske, F.; Birner, P. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) correlates with Her-2 status, carbonic anhydrase 9 expression and prognosis in esophageal cancer. Clin. Exp. Metast. 2012, 29, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.F.; Chen, P.T.; Lu, M.S.; Lin, P.Y.; Chen, W.C.; Lee, K.D. IL-6 expression predicts treatment response and outcome in squamous cell carcinoma of the esophagus. Mol. Cancer 2013, 12, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.Y.; Yu, J.; Liu, T.T.; Yang, K.X.; Yang, L.Y.; Chen, Q.; Shi, F.; Hao, J.J.; Cai, Y.; Wang, M.R.; et al. Plumbagin inhibits the proliferation and survival of esophageal cancer cells by blocking STAT3-PLK1-AKT signaling. Cell Death Dis. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Li, S.; Duan, X.; Gu, Z.; Ma, Z.; Yuan, X.; Feng, X.; Wang, H. Inhibition of glycogen synthase kinase 3 beta (GSK3beta) suppresses the progression of esophageal squamous cell carcinoma by modifying STAT3 activity. Mol. Carcinog. 2017, 56, 2301–2316. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.F.; Zhang, Z.T.; Wang, J.Y.; Xu, B.B. Icariin exerts inhibitory effects on the growth and metastasis of KYSE70 human esophageal carcinoma cells via PI3K/AKT and STAT3 pathways. Environ. Toxicol. Pharmacol. 2017, 54, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Xuan, X.; Li, S.; Lou, X.; Zheng, X.; Li, Y.; Wang, F.; Gao, Y.; Zhang, H.; He, H.; Zeng, Q. Stat3 promotes invasion of esophageal squamous cell carcinoma through up-regulation of MMP2. Mol. Biol. Rep. 2015, 42, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Rice, T.W.; Blackstone, E.H.; Rusch, V.W. 7th edition of the AJCC Cancer Staging Manual: Esophagus and esophagogastric junction. Ann. Surg. Oncol. 2010, 17, 1721–1724. [Google Scholar] [CrossRef] [PubMed]
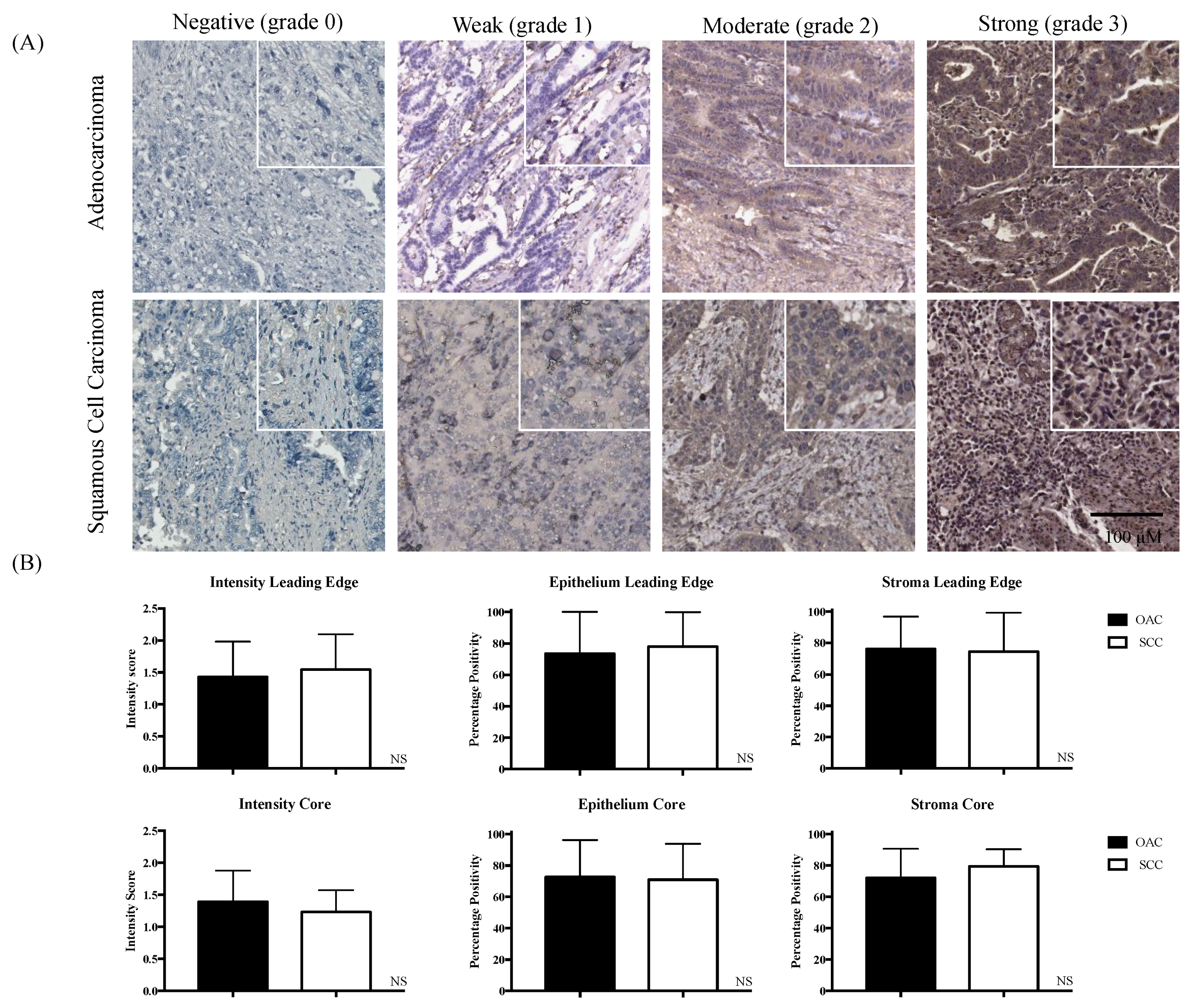
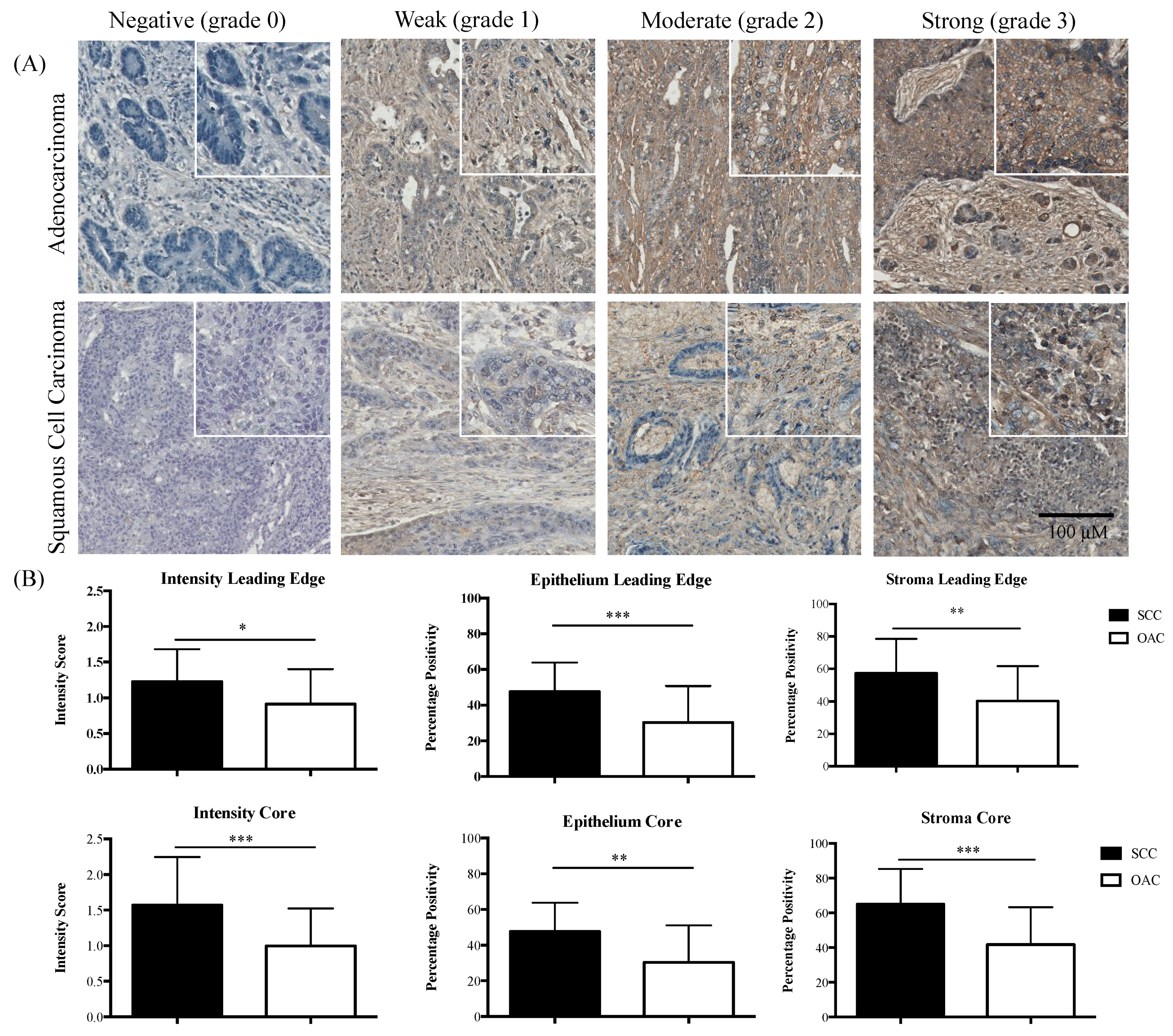


| Core | Leading Edge | |||||
|---|---|---|---|---|---|---|
| Adenocarcinoma | ||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Intensity | 1.107 | 0.661, 1.856 | 0.699 | 0.449 | 0.194, 1.037 | 0.061 |
| Epithelium | 1.065 | 0.843, 1.345 | 0.600 | 0.740 | 0.574, 0.953 | 0.020 |
| Stroma | 1.192 | 0.890, 1.598 | 0.239 | 0.660 | 0.425, 1.025 | 0.064 |
| Squamous Cell Carcinoma | ||||||
| Intensity | 1.164 | 0.288, 4.701 | 0.831 | 1.514 | 0.327, 7.015 | 0.596 |
| Epithelium | 1.189 | 0.768, 1.839 | 0.437 | 3.726 | 0.871, 15.938 | 0.076 |
| Stroma | 6.382 | 1.266, 32.184 | 0.025 | 1.222 | 0.419, 3.570 | 0.713 |
| Adenocarcinoma (n = 116) | Squamous Cell Carcinoma (n = 38) | Total (n = 154) | ||
|---|---|---|---|---|
| Age | Mean ± sd | 64.53 ± 11.75 | 63.63 ± 11.4 | 64.48 ± 11.18 |
| BMI | Mean ± sd | 26.3 ± 4.5 (88) | 23.4 ± 4.2 (30) | 25.6 ± 4.6 (118) |
| Involvement n (%) | ||||
| Lymph node | 79 (69.3) | 21 (56.8) | 100 (66.2) | |
| Venous | 55 (49.5) | 15 (40.5) | 70 (47.3) | |
| Perineural | 50 (43.9) | 8 (21.6) | 58 (38.4) | |
| Involved resection margins | 62 (54.4) | 25 (65.8) | 87 (57.2) | |
| Differentiation | ||||
| Well | 6 (5.2) | 5 (13.2) | 11 (7.1) | |
| Moderate | 67 (57.8) | 24 (63.2) | 91 (59.1) | |
| Poor | 43 (37.1) | 9 (23.7) | 52 (33.8) | |
| Microscopic residual tumour | 29 (25.9) | 8 (21.6) | 37 (24.8) | |
| Staging n (%) | ||||
| T Stage | In situ | 0 | 1 (2.7) | 1 (0.7) |
| T1 | 14 (12.2) | 3 (8.1) | 17 (11.2) | |
| T2 | 20 (17.4) | 7 (18.9) | 27 (17.8) | |
| T3 | 77 (67.0) | 25 (67.6) | 102 (67.1) | |
| T4 | 4 (3.5) | 1 (2.7) | 5 (3.3) | |
| N stage | N0 | 39 (33.9) | 14 (37.8) | 53 (34.9) |
| N1 | 62 (53.9) | 23 (62.2) | 85 (55.9) | |
| N2 | 9 (7.8) | 0 | 9 (5.9) | |
| N3 | 5 (4.3) | 0 | 5 (3.3) | |
| M stage | M1 | 3 (18.8) | 2 (40.0) | 5 (23.8) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’ Sullivan, K.E.; Michielsen, A.J.; O’ Regan, E.; Cathcart, M.C.; Moore, G.; Breen, E.; Segurado, R.; Reynolds, J.V.; Lysaght, J.; O’ Sullivan, J. pSTAT3 Levels Have Divergent Expression Patterns and Associations with Survival in Squamous Cell Carcinoma and Adenocarcinoma of the Oesophagus. Int. J. Mol. Sci. 2018, 19, 1720. https://doi.org/10.3390/ijms19061720
O’ Sullivan KE, Michielsen AJ, O’ Regan E, Cathcart MC, Moore G, Breen E, Segurado R, Reynolds JV, Lysaght J, O’ Sullivan J. pSTAT3 Levels Have Divergent Expression Patterns and Associations with Survival in Squamous Cell Carcinoma and Adenocarcinoma of the Oesophagus. International Journal of Molecular Sciences. 2018; 19(6):1720. https://doi.org/10.3390/ijms19061720
Chicago/Turabian StyleO’ Sullivan, Katie E., Adriana J. Michielsen, Esther O’ Regan, Mary C. Cathcart, Gillian Moore, Eamon Breen, Ricardo Segurado, John V. Reynolds, Joanne Lysaght, and Jacintha O’ Sullivan. 2018. "pSTAT3 Levels Have Divergent Expression Patterns and Associations with Survival in Squamous Cell Carcinoma and Adenocarcinoma of the Oesophagus" International Journal of Molecular Sciences 19, no. 6: 1720. https://doi.org/10.3390/ijms19061720




