Circulating MicroRNAs in Young Patients with Acute Coronary Syndrome
Abstract
:1. Introduction
2. Results
2.1. Baseline Characteristics of the Study Population
2.2. Detection of miRNA Expression in Plasma of Young ACS by Small RNA Sequencing (sRNA-seq)
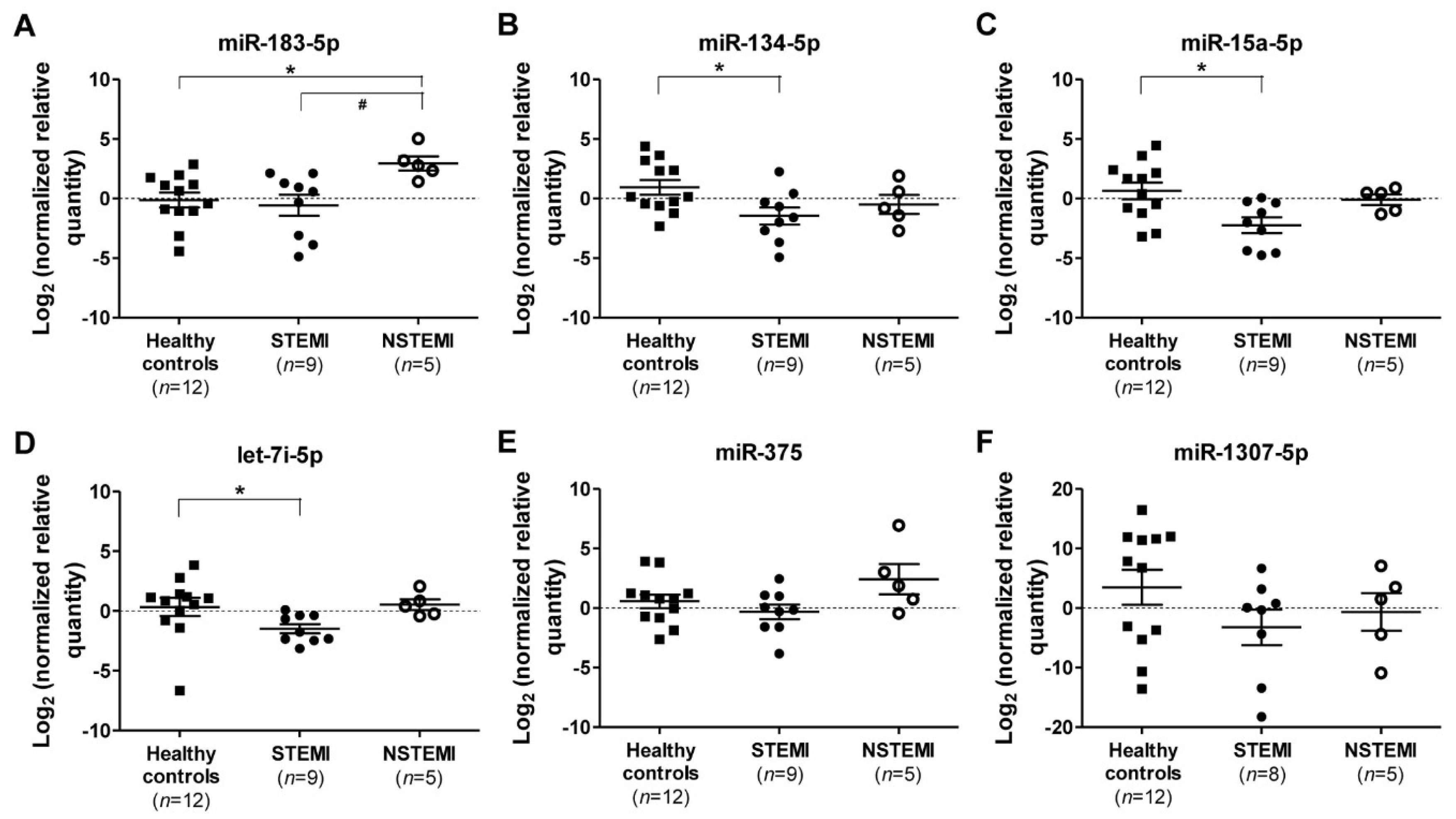
2.3. Quantitative RT-PCR (qRT-PCR) Validation of miRNAs Expression
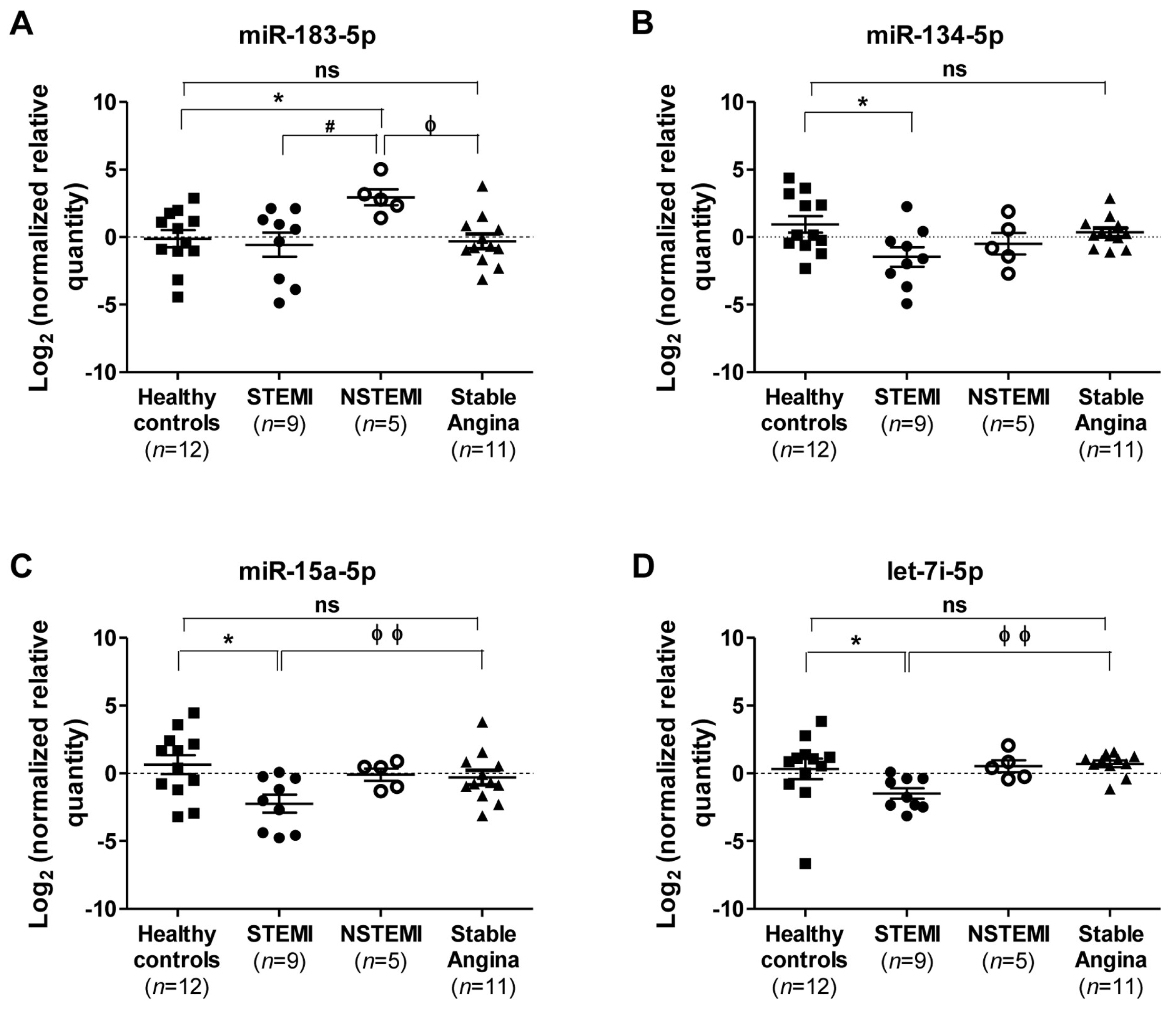
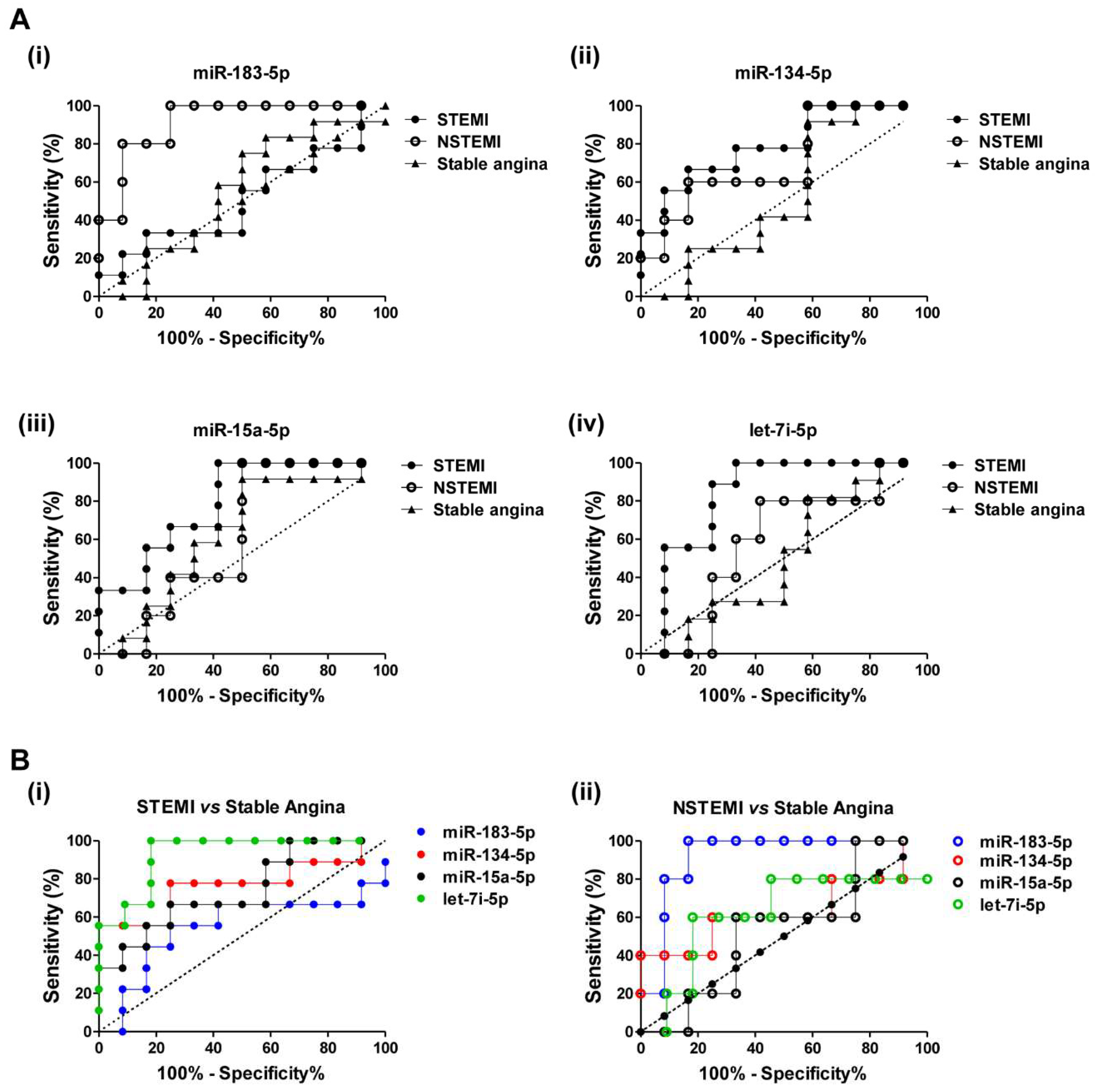
2.4. Diagnostic Power of Plasma miRNAs in STEMI and NSTEMI Patients
2.5. Discriminatory Power of the miRNA Combination Panel
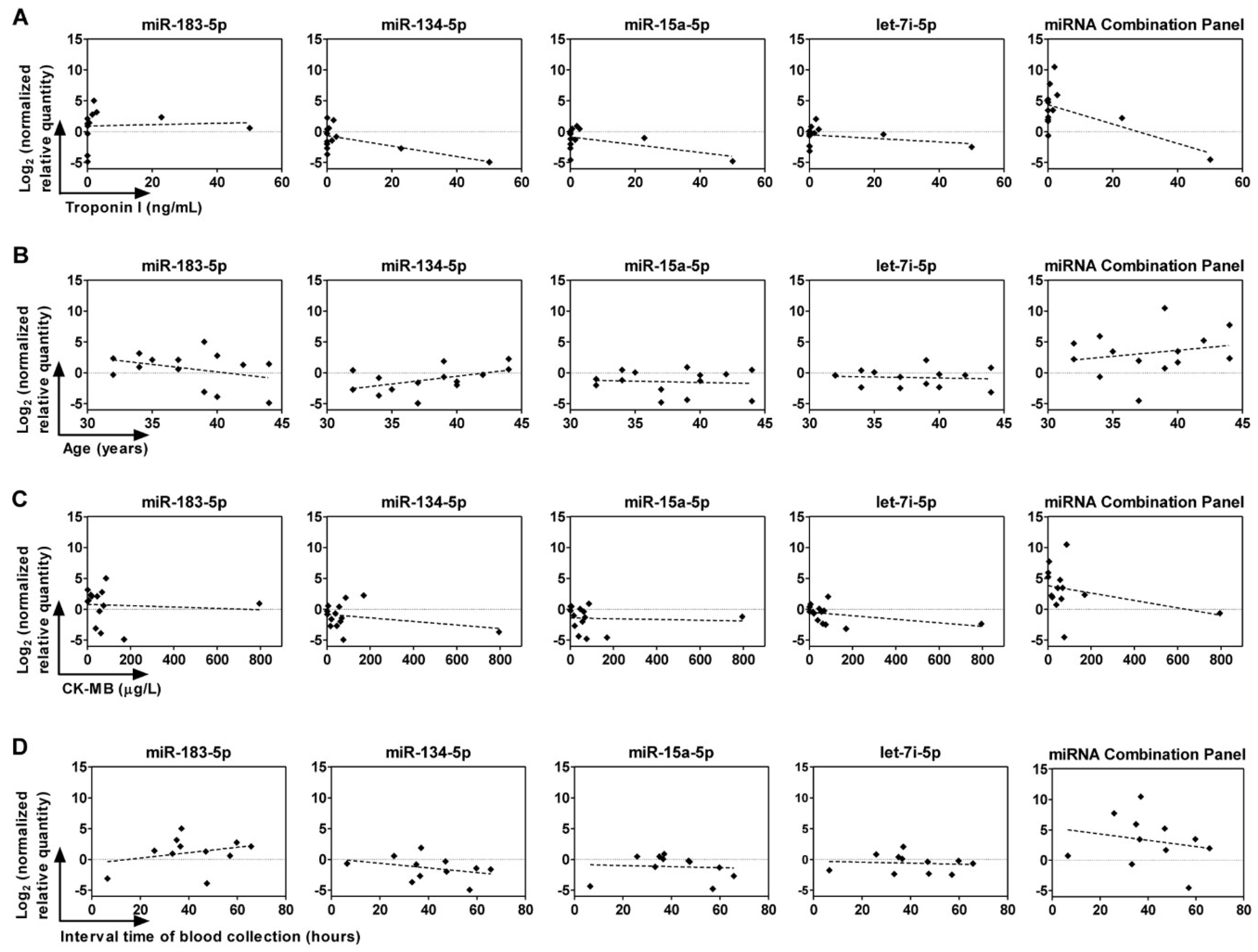
2.6. Correlation between Circulating miRNAs, Age, and Troponin I and CK-MB Levels
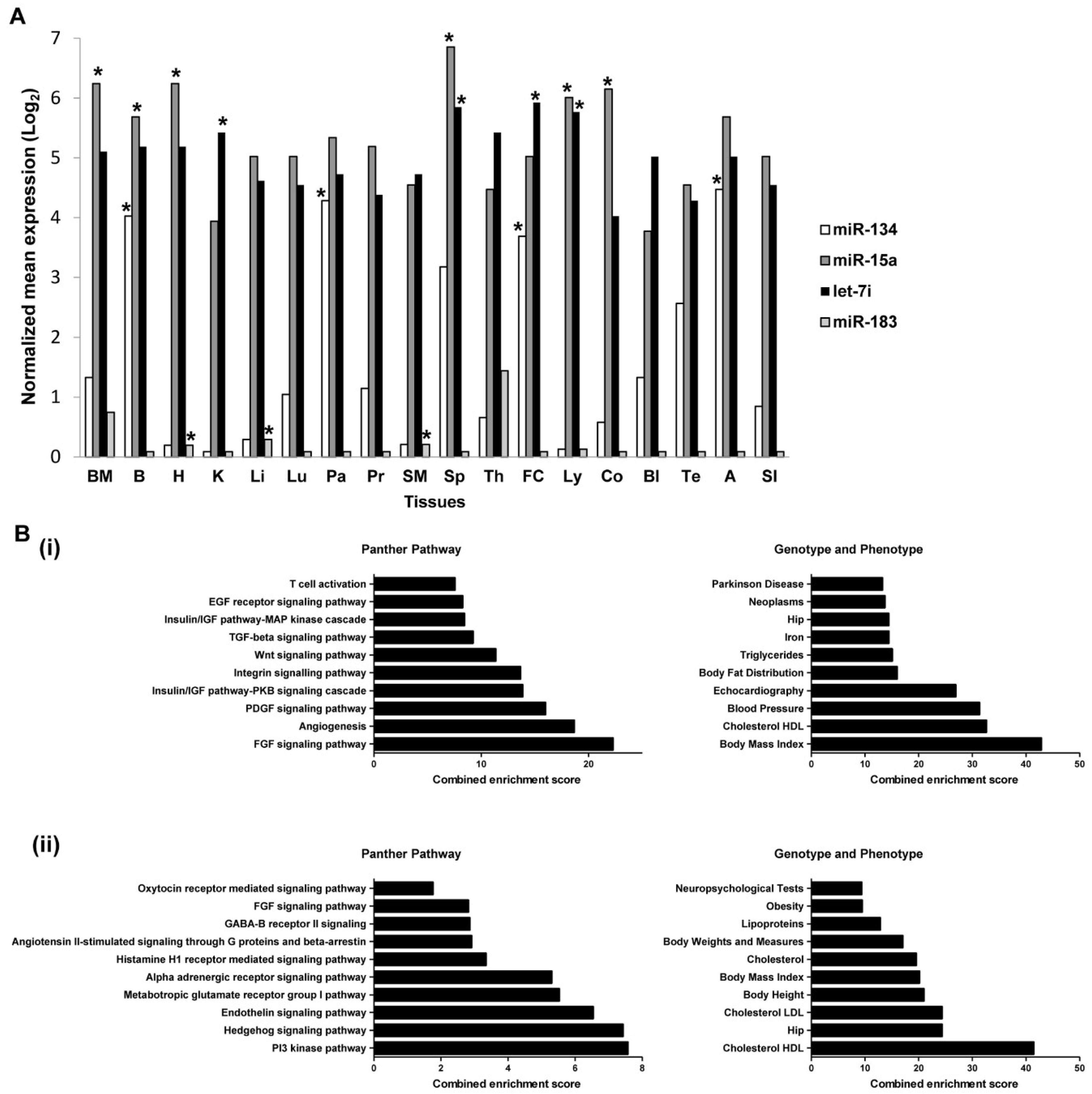
2.7. Expressions of miRNAs in Tissues and Organs
2.8. Predicted Target Genes of miRNAs Are Enriched in Metabolic Pathways
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Isolation of Human Plasma
4.3. MiRNA Extraction
4.4. Small-RNA Sequencing (sRNA-seq)
4.5. Sequencing Data Analysis
4.6. Validation of miRNA by qRT-PCR
4.7. Tissue Expression Analysis of miRNA Candidates
4.8. Target Genes Prediction and Gene List Enrichment Analysis for miRNA Candidates
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| CAD | Coronary artery disease |
| ACS | Acute coronary syndrome |
| STEMI | ST-segment elevated myocardial infarction |
| NSTEMI | Non-ST-segment elevated myocardial infarction |
| ECG | Electrocardiography |
| CK-MB | Creatine kinase-myocardial band |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| HDL | High-density lipoproteins |
| sRNA-seq | Small RNA sequencing |
| FDR | False discovery rate |
| ANOVA | Analysis of variance |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| 95% CI | 95% confidence interval |
| mESAdb | microRNA Expression and Sequence Analysis database |
| LDL | Low-density lipoproteins |
| VSMC | Vascular smooth muscle cells |
| SRs | Scavenger receptors |
| AGTR1 | Angiotensin II type 1 receptor |
| CVD | Cardiovascular disease |
| EDTA | Ethylenediaminetetraacetic acid |
| NRQ | Normalized relative quantity |
| RQ | Relative quantity |
| NF | Normalizing factor |
References
- Cassar, A.; Holmes, D.R.; Rihal, C.S.; Gersh, B.J. Chronic Coronary Artery Disease: Diagnosis and Management. Mayo Clin. Proc. 2009, 84, 1130–1146. [Google Scholar] [CrossRef] [PubMed]
- Morrow, D.A.; Cannon, C.P.; Jesse, R.L.; Newby, L.K.; Ravkilde, J.; Storrow, A.B.; Wu, A.H.; Christenson, R.H. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines: Clinical characteristics and utilization of biochemical markers in acute coronary syndromes. Circulation 2007, 115, e356–e375. [Google Scholar] [CrossRef] [PubMed]
- Pollehn, T.; Brady, W.J.; Perron, A.D.; Morris, F. The electrocardiographic differential diagnosis of ST segment depression. Emerg. Med. J. 2002, 19, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Brady, W.J.; Roberts, D.; Morris, F. The nondiagnostic ECG in the chest pain patient: Normal and nonspecific initial ECG presentations of acute MI. Am. J. Emerg. Med. 1999, 17, 394–397. [Google Scholar] [CrossRef]
- Venge, P.; James, S.; Jansson, L.; Lindahl, B. Clinical performance of two highly sensitive cardiac troponin I assays. Clin. Chem. 2009, 55, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Dekker, M.S.; Mosterd, A.; van Hof, A.W.; Hoes, A.W. Novel biochemical markers in suspected acute coronary syndrome: Systematic review and critical appraisal. Heart 2010, 96, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Tanindi, A.; Cemri, M. Troponin elevation in conditions other than acute coronary syndromes. Vasc. Health Risk Manag. 2011, 7, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Van Empel, V.P.; De Windt, L.J.; da Costa Martins, P.A. Circulating miRNAs: Reflecting or affecting cardiovascular disease? Curr. Hypertens. Rep. 2012, 14, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Wronska, A.; Kurkowska-Jastrzebska, I.; Santulli, G. Application of microRNAs in diagnosis and treatment of cardiovascular disease. Acta Physiol. 2015, 213, 60–83. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Touvana, E.; Stefanou, C.; Deltas, C. microRNAs: A newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010, 14, 236–240. [Google Scholar] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef] [PubMed]
- Ganci, F.; Sacconi, A.; Manciocco, V.; Covello, R.; Benevolo, M.; Rollo, F.; Strano, S.; Valsoni, S.; Bicciato, S.; Spriano, G.; et al. Altered peritumoral microRNA expression predicts head and neck cancer patients with a high risk of recurrence. Mod. Pathol. 2017, 30, 1387. [Google Scholar] [CrossRef] [PubMed]
- Pennati, M.; Folini, M.; Gandellini, P.; Zaffaroni, N. MicroRNAs and the Response of Prostate Cancer to Anti-Cancer Drugs. Curr. Drug Targets 2016, 17, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Andreou, I.; Sun, X.; Stone, P.H.; Edelman, E.R.; Feinberg, M.W. miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends Mol. Med. 2015, 21, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Bronze-da-Rocha, E. MicroRNAs expression profiles in cardiovascular diseases. BioMed Res. Int. 2014, 2014, 985408. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.; van der Lans, C.A.; Halvorsen, B.; Gullestad, L.; Kuiper, J.; Aukrust, P.; van Berkel, T.J.; Biessen, E.A. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem. Biophys. Res. Commun. 2010, 394, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jing, Z.C.; Ellinor, P.T.; Liang, D.; Zhang, H.; Liu, Y.; Chen, X.; Pan, L.; Lyon, R.; Liu, Y.; et al. MicroRNA-134 as a potential plasma biomarker for the diagnosis of acute pulmonary embolism. J. Transl. Med. 2011, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Oerlemans, M.I.; Mosterd, A.; Dekker, M.S.; de Vrey, E.A.; van Mil, A.; Pasterkamp, G.; Doevendans, P.A.; Hoes, A.W.; Sluijter, J.P. Early assessment of acute coronary syndromes in the emergency department: The potential diagnostic value of circulating microRNAs. EMBO Mol. Med. 2012, 4, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Zhao, X.; Liu, Y.Z.; Zeng, Q.T.; Mao, X.B.; Li, S.N.; Zhang, M.; Jiang, C.; Zhou, Y.; Qian, C.; et al. Circulating MiR-19b-3p, MiR-134-5p and MiR-186-5p are Promising Novel Biomarkers for Early Diagnosis of Acute Myocardial Infarction. Cell. Physiol. Biochem. 2016, 38, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Busby-Whitehead, J.; Alexander, K.P. Acute coronary syndrome in the older adults. J. Geriatr. Cardiol. JGC 2016, 13, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gacon, J.; Kablak-Ziembicka, A.; Stepien, E.; Enguita, F.J.; Karch, I.; Derlaga, B.; Zmudka, K.; Przewlocki, T. Decision-making microRNAs (miR-124, -133a/b, -34a and -134) in patients with occluded target vessel in acute coronary syndrome. Kardiol. Polska 2016, 74, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Cantu-Brito, C.; Chiquete, E.; Ruiz-Sandoval, J.L.; Gaxiola, E.; Albuquerque, D.C.; Corbalan, R.; Ramos, A.; Bhatt, D.L.; Steg, P.G. Atherothrombotic disease, traditional risk factors, and 4-year mortality in a Latin American population: The REACH Registry. Clin. Cardiol. 2012, 35, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Previtali, E.; Bucciarelli, P.; Passamonti, S.M.; Martinelli, I. Risk factors for venous and arterial thrombosis. Blood Transfus. 2011, 9, 120–138. [Google Scholar] [PubMed]
- Wan Azman, W.A.; Sim, K.H. Annual Report of the NCVD-ACS Registry, Year 2011–2013; National Heart Association of Malaysia (NHAM) and the Clinical Research Centre, Ministry of Health Malaysia: Kuala Lumpur, Malaysia, 2015. [Google Scholar]
- Muda, Z.; Kadir, A.A.; Yusof, Z.; Yaacob, L.H. Premature Coronary Artery Disease among Angiographically Proven Atherosclerotic Coronary Artery Disease in North East of Peninsular Malaysia. Int. J. Collab. Res. Intern. Med. Public Health 2013, 5, 507–516. [Google Scholar]
- Arantes, C.; Martins, J.; Braga, C.G.; Ramos, V.; Vieira, C.; Gaspar, A.; Salgado, A.; Pereira, M.A.; Rocha, S.; Correia, A. Acute coronary syndrome in young adults. Eur. Heart J. 2013, 34 (Suppl. 1), P3134. [Google Scholar] [CrossRef]
- Teixeira, M.; Sa, I.; Mendes, J.S.; Martins, L. Acute coronary syndrome in young adults. Port. J. Cardiol. Off. J. Port. Soc. Cardiol. 2010, 29, 947–955. [Google Scholar]
- Che, J.; Li, G.; Shao, Y.; Niu, H.; Shi, Y. An analysis of the risk factors for premature coronary artery disease in young and middle-age Chinese patients with hypertension. Exp. Clin. Cardiol. 2013, 18, 89–92. [Google Scholar] [PubMed]
- Kang, S.M.; Choi, J.W.; Lee, Y.; Hong, S.H.; Lee, H.J. Identification of microRNA-size, small RNAs in Escherichia coli. Curr. Microbiol. 2013, 67, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Kaya, K.D.; Karakulah, G.; Yakicier, C.M.; Acar, A.C.; Konu, O. mESAdb: MicroRNA expression and sequence analysis database. Nucleic Acids Res. 2011, 39, D170–D180. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Economou, E.K.; Oikonomou, E.; Siasos, G.; Papageorgiou, N.; Tsalamandris, S.; Mourouzis, K.; Papaioanou, S.; Tousoulis, D. The role of microRNAs in coronary artery disease: From pathophysiology to diagnosis and treatment. Atherosclerosis 2015, 241, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Jones Buie, J.N.; Goodwin, A.J.; Cook, J.A.; Halushka, P.V.; Fan, H. The role of miRNAs in cardiovascular disease risk factors. Atherosclerosis 2016, 254, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Navickas, R.; Gal, D.; Laucevičius, A.; Taparauskaitė, A.; Zdanytė, M.; Holvoet, P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: A systematic review. Cardiovasc. Res. 2016, 111, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Widera, C.; Gupta, S.K.; Lorenzen, J.M.; Bang, C.; Bauersachs, J.; Bethmann, K.; Kempf, T.; Wollert, K.C.; Thum, T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J. Mol. Cell. Cardiol. 2011, 51, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Karakas, M.; Schulte, C.; Appelbaum, S.; Ojeda, F.; Lackner, K.J.; Munzel, T.; Schnabel, R.B.; Blankenberg, S.; Zeller, T. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur. Heart J. 2017, 38, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Adachi, T.; Nakanishi, M.; Otsuka, Y.; Nishimura, K.; Hirokawa, G.; Goto, Y.; Nonogi, H.; Iwai, N. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin. Chem. 2010, 56, 1183–1185. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Zhang, R.; Li, Y.; Pu, J.; Lu, Y.; Jiao, J.; Li, K.; Yu, B.; Li, Z.; Wang, R.; et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem. Biophys. Res. Commun. 2010, 391, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Tan, N.; Yang, J.; Liu, X.; Cao, X.; He, P.; Dong, X.; Qin, S.; Zhang, C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin. Sci. 2010, 119, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.R.; Staessen, J.A.; Heymans, S.; Schroen, B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 499–506. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandra, Y.; Devanna, P.; Limana, F.; Straino, S.; Di Carlo, A.; Brambilla, P.G.; Rubino, M.; Carena, M.C.; Spazzafumo, L.; De Simone, M.; et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 2010, 31, 2765–2773. [Google Scholar] [CrossRef] [PubMed]
- Gidlof, O.; Andersson, P.; van der Pals, J.; Gotberg, M.; Erlinge, D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology 2011, 118, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Takahashi, R.; Hiura, Y.; Hirokawa, G.; Fukushima, Y.; Iwai, N. Plasma miR-208 as a biomarker of myocardial injury. Clin. Chem. 2009, 55, 1944–1949. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, Y.; Ono, K.; Horie, T.; Nishi, H.; Nagao, K.; Kinoshita, M.; Watanabe, S.; Baba, O.; Kojima, Y.; Shizuta, S.; et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ. Cardiovasc. Genet. 2011, 4, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Antonicelli, R.; Lorenzi, M.; D’Alessandra, Y.; Lazzarini, R.; Santini, G.; Spazzafumo, L.; Lisa, R.; La Sala, L.; Galeazzi, R.; et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int. J. Cardiol. 2013, 167, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.K.; Zhu, J.Q.; Zhang, J.T.; Li, Q.; Li, Y.; He, J.; Qin, Y.W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.F.; Neylon, A.; McGorrian, C.; Blake, G.J. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 2016, 224, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Minami, Y.; Takahashi, Y.; Tabuchi, T.; Nakamura, M. A cellular microRNA, let-7i, is a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. J. Card. Fail. 2011, 17, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.C.; Wang, Y.S.; Guo, Y.C.; Lin, W.L.; Chang, M.H.; Juo, S.H. Let-7g improves multiple endothelial functions through targeting transforming growth factor-beta and SIRT-1 signaling. J. Am. Coll. Cardiol. 2014, 63, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Lv, P.; Zhao, X.; Wang, X.; Ma, X.; Meng, W.; Meng, X.; Dong, S. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol. Cell. Biochem. 2014, 394, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Lan, G.; Xie, W.; Li, L.; Zhang, M.; Liu, D.; Tan, Y.L.; Cheng, H.P.; Gong, D.; Huang, C.; Zheng, X.L.; et al. MicroRNA-134 actives lipoprotein lipase-mediated lipid accumulation and inflammatory response by targeting angiopoietin-like 4 in THP-1 macrophages. Biochem. Biophys. Res. Commun. 2016, 472, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Condorelli, G.; Mehta, J.L. Scavenger receptors and non-coding RNAs: Relevance in atherogenesis. Cardiovasc. Res. 2016, 109, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Gavard, J.; Gutkind, J.S. VE-cadherin and claudin-5: It takes two to tango. Nat. Cell Biol. 2008, 10, 883–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.W.; Wu, H.C.; Terry, M.B.; Santella, R.M. microRNA Expression in Prospectively Collected Blood as a Potential Biomarker of Breast Cancer Risk in the BCFR. Anticancer Res. 2015, 35, 3969–3977. [Google Scholar] [PubMed]
- Tang, J.F.; Yu, Z.H.; Liu, T.; Lin, Z.Y.; Wang, Y.H.; Yang, L.W.; He, H.J.; Cao, J.; Huang, H.L.; Liu, G. Five miRNAs as novel diagnostic biomarker candidates for primary nasopharyngeal carcinoma. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 7575–7581. [Google Scholar] [CrossRef] [PubMed]
- Wojcicka, A.; Swierniak, M.; Kornasiewicz, O.; Gierlikowski, W.; Maciag, M.; Kolanowska, M.; Kotlarek, M.; Gornicka, B.; Koperski, L.; Niewinski, G.; et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2014, 53, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xiang, G.; Meng, Y.; Dong, R. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod. Biol. 2016, 16, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Krug, S.M.; Heitmann, J.; Hu, L.; Reinhold, A.K.; Sauer, S.; Bosten, J.; Sommer, C.; Fromm, M.; Brack, A.; et al. Analgesic drug delivery via recombinant tissue plasminogen activator and microRNA-183-triggered opening of the blood-nerve barrier. Biomaterials 2016, 82, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Sasaki, H.; Furuse, M.; Tsukita, S. Endothelial Claudin: Claudin-5/Tmvcf Constitutes Tight Junction Strands in Endothelial Cells. J. Cell Biol. 1999, 147, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.L.; Armugam, A.; Sepramaniam, S.; Karolina, D.S.; Lim, K.Y.; Lim, J.Y.; Chong, J.P.; Ng, J.Y.; Chen, Y.T.; Chan, M.M.; et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur. J. Heart Fail. 2015, 17, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Li, S.; Li, J.; Yin, L.; Zhou, T.; Zhang, C.; Chen, X.; Sun, K. Ethnic differences in microRNA-375 expression level and DNA methylation status in type 2 diabetes of Han and Kazak populations. J. Diabetes Res. 2014, 2014, 761938. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.; Bagnall, R.D.; Peverill, R.E.; Donelan, L.; Corben, L.; Delatycki, M.B.; Semsarian, C. A polymorphic miR-155 binding site in AGTR1 is associated with cardiac hypertrophy in Friedreich ataxia. J. Mol. Cell. Cardiol. 2011, 51, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, P.; Borel, C.; Gagnebin, M.; Grant, G.R.; Deutsch, S.; Elton, T.S.; Hatzigeorgiou, A.G.; Antonarakis, S.E. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3’ untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am. J. Hum. Genet. 2007, 81, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.X.; Zeng, D.Y.; Li, R.T.; Pang, R.P.; Yang, H.; Hu, Y.L.; Zhang, Q.; Jiang, Y.; Huang, L.Y.; Tang, Y.B.; et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 2012, 60, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cao, C.; Chen, H.; Song, J.; Lee, C.; Zhang, J.; Zhang, F.; Geng, Q.; Li, Z.; Li, J. Atheroprotective effects of statins in patients with unstable angina by regulating the blood-borne microRNA network. Mol. Med. Rep. 2017, 16, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health. Clinical Practice Guidelines on Management of Unstable Angina/Non ST Elevation Myocardial Infarction (UA/NSTEMI); Ministry of Health: Putrajaya, Malaysia, 2011.
- Ministry of Health Malaysia. Clinical Practice Guidelines—Management of Acute ST Segment Elevation Myocardial Infarction (STEMI), 3rd ed.; Ministry of Health Malaysia: Putrajaya, Malaysia, 2014.
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Royston, P. A Remark on Algorithm AS 181: The W-test for Normality. J. R. Stat. Soc. Ser. C 1995, 44, 547–551. [Google Scholar]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 3–4. [Google Scholar] [CrossRef]
- Sheskin, D.J. Handbook of Parametric and Nonparametric Statistical Procedures, 5th ed.; Chapman & Hall/CRC 2000: Boca Raton, FL, USA, 2011. [Google Scholar]
- Thode, H.J. Testing for Normality; Marcel Dekker: New York, NY, USA, 2002. [Google Scholar]
- Liu, W.; Ling, S.; Sun, W.; Liu, T.; Li, Y.; Zhong, G.; Zhao, D.; Zhang, P.; Song, J.; Jin, X.; et al. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci. Rep. 2015, 5, 16099. [Google Scholar] [CrossRef] [PubMed]
- Goren, Y.; Kushnir, M.; Zafrir, B.; Tabak, S.; Lewis, B.S.; Amir, O. Serum levels of microRNAs in patients with heart failure. Eur. J. Heart Fail. 2012, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]


| STEMI Patients (n = 11) | NSTEMI Patients (n = 3) | Healthy Controls (n = 14) | p-Value | |
|---|---|---|---|---|
| Age (years) | 36.36 ± 1.73 | 39.67 ± 2.96 | 35.55 ± 1.92 | 0.5851 |
| Male sex (% male) | 11 (100) | 3 (100) | 14 (100) | |
| Health history, n (%): | ||||
| Alcohol drinker | 1 (9.09) | 0 (0) | 0 (0) | |
| Smokers | 11 (100) | 1 (33.33) | 0 (0) | |
| Morbid Obesity | 0 (0) | 0 (0) | 0 (0) | |
| Familial Hypercholesterolemia | 1 (9.09) | 0 (0) | 0 (0) | |
| Hypertension | 4 (36.36) | 0 (0) | 1 (7.14) | |
| Diabetes mellitus | 1 (9.09) | 1 (33.33) | 1 (7.14) | |
| Family History of CVD | 7 (63.64) | 2 (66.67) | 0 (0) | |
| # Medications, n (%): | ||||
| Acetylsalicylic acid | 9 (81.82) | 2 (66.67) | 0 (0) | |
| Lipid lowering drugs | 10 (90.91) | 2 (66.67) | 0 (0) | |
| Beta-blocker | 6 (54.55) | 1 (33.33) | 0 (0) | |
| Others | 2 (18.18) | 1 (33.33) | 0 (0) | |
| Laboratory tests | ||||
| WBC (×109/L) | 12.63 ± 1.14 | 11.90 ± 1.79 | 8.68 ± 0.56 | 0.0110 * |
| Glucose (mmol/L) | 6.19 ± 0.55 | 8.10 ± 2.61 | 6.26 ± 0.58 | 0.4087 |
| TC (mmol/L) | 4.83 ± 0.40 | 5.07 ± 0.45 | 5.26 ± 0.44 | 0.7581 |
| HDL (mmol/L) | 0.96 ± 0.07 | 0.78 ± 0.10 | 1.19 ± 0.09 | 0.0329 * |
| LDL (mmol/L) | 3.14 ± 0.37 | 3.39 ± 0.30 | 3.33 ± 0.38 | 0.5610 |
| TG (mmol/L) | 1.61 ± 0.22 | 1.97 ± 0.52 | 1.65 ± 0.20 | 0.7493 |
| Troponin I (ng/mL) | 11.78 ± 4.89 | 1.94 ± 0.69 | ND | ND |
| Creatine kinase (U/L) | 1859.67 ± 619.44 | 496.00 ± 337.93 | 641.3 ± 495.96 | 0.1037 |
| CK-MB (µg/L) | 88.33 ± 25.69 | 28.00 ± 25.03 | 4.03 ± 2.07 | 0.0698 |
| STEMI Patients (n = 9) | NSTEMI Patients (n = 5) | Healthy Controls (n = 12) | p-Value | |
|---|---|---|---|---|
| Age (years) | 37.78 ± 1.29 | 37.80 ± 2.15 | 36.80 ± 1.83 | 0.8912 |
| Male sex (% male) | 9 (100) | 5 (100) | 12 (100) | |
| Health history, n (%): | ||||
| Alcohol drinker | 1 (11.11) | 0 (0) | 0 (0) | |
| Smokers | 8 (88.89) | 2 (40.00) | 0 (0) | |
| Morbid Obesity | 0 (0) | 0 (0) | 0 (0) | |
| Familial Hypercholesterolemia | 0 (0) | 0 (0) | 0 (0) | |
| Hypertension | 0 (0) | 0 (0) | 1 (8.33) | |
| Diabetes mellitus | 0 (0) | 2 (40.00) | 1 (8.33) | |
| Family History of CVD | 5 (55.56) | 3 (60.00) | 0 (0) | |
| # Medications, n (%): | ||||
| Acetylsalicylic acid | 8 (88.89) | 3 (60.00) | 0 (0) | |
| Lipid lowering drugs | 8 (88.89) | 3 (60.00) | 0 (0) | |
| Beta-blocker | 7 (77.78) | 2 (40.00) | 0 (0) | |
| Others | 1 (11.11) | 1 (20.00) | 0 (0) | |
| Laboratory tests | ||||
| WBC (×109/L) | 13.22 ± 1.63 | 12.02 ± 1.17 | 9.13 ± 0.74 | 0.0587 |
| Glucose (mmol/L) | 6.33 ± 0.49 | 9.04 ± 2.00 | 7.10 ± 0.71 | 0.1979 |
| TC (mmol/L) | 5.66 ± 0.62 | 5.46 ± 0.41 | 5.62 ± 0.49 | 0.9685 |
| HDL (mmol/L) | 1.14 ± 0.26 | 0.99 ± 0.12 | 1.27 ± 0.09 | 0.0841 |
| LDL (mmol/L) | 3.55 ± 0.68 | 3.80 ± 0.37 | 3.58 ± 0.46 | 0.7561 |
| TG (mmol/L) | 2.04 ± 0.29 | 1.46 ± 0.16 | 1.72 ± 0.27 | 0.9510 |
| Troponin I (ng/mL) | 6.29 ± 6.24 | 5.97 ± 4.23 | ND | ND |
| Creatine kinase (U/L) | 1526.89 ± 461.82 | 760.20 ± 452.37 | 910.50 ± 721.50 | 0.3601 |
| CK-MB (µg/L) | 140.28 ± 83.35 | 35.20 ± 17.47 | 5.55 ± 2.45 | 0.2262 |
| miRNA | Base Mean | Log2 Fold Change | Fold Change | Regulation | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|---|
| hsa-miR-183-5p_MIMAT0000261 | 133.430 | 2.232 | 4.699 | Up | 0.0000856 | 0.00317 |
| hsa-miR-19a-5p_MIMAT0004490 | 269.760 | 1.732 | 3.322 | Up | 0.00379 | 0.03740 |
| hsa-miR-15a-5p_MIMAT0000068 | 466.668 | 1.680 | 3.205 | Up | 0.00252 | 0.02723 |
| hsa-miR-101-5p_MIMAT0004513 | 1857.448 | 1.053 | 2.075 | Up | 0.00942 | 0.07019 |
| hsa-miR-101-3p_MIMAT0000099 | 1857.448 | 1.053 | 2.075 | Up | 0.00942 | 0.07019 |
| hsa-miR-103a-3p_MIMAT0000101 | 5231.824 | 1.047 | 2.066 | Up | 0.00250 | 0.02723 |
| hsa-miR-107_MIMAT0000104 | 5231.824 | 1.047 | 2.066 | Up | 0.00250 | 0.02723 |
| hsa-miR-103a-2-5p_MIMAT0009196 | 5231.824 | 1.047 | 2.066 | Up | 0.00250 | 0.02723 |
| hsa-miR-16-5p_MIMAT0000069 | 5923.048 | 1.037 | 2.052 | Up | 0.01173 | 0.07091 |
| hsa-miR-16-5p_MIMAT0000069 | 5925.776 | 1.036 | 2.051 | Up | 0.01180 | 0.07091 |
| hsa-miR-25-5p_MIMAT0004498 | 18,099.042 | 0.918 | 1.890 | Up | 0.00473 | 0.04118 |
| hsa-miR-30e-5p_MIMAT0000692 | 2296.889 | 0.897 | 1.862 | Up | 0.01612 | 0.08520 |
| hsa-miR-30a-5p_MIMAT0000087 | 3027.454 | 0.849 | 1.801 | Up | 0.01892 | 0.09657 |
| CM000680.1_15285_novel | 3128.361 | −2.349 | 5.093 | Down | 0.0000004213 | 0.00006 |
| CM000668.1_36492_novel | 1075.400 | −2.202 | 4.602 | Down | 0.00122 | 0.02578 |
| hsa-miR-1307-5p_MIMAT0022727 | 404.613 | −2.191 | 4.565 | Down | 0.00003364 | 0.00166 |
| hsa-miR-375_MIMAT0000728 | 1029.877 | −2.058 | 4.164 | Down | 0.00036 | 0.01060 |
| hsa-let-7i-5p_MIMAT0000415 | 58,413.753 | −1.910 | 3.759 | Down | 0.000009499 | 0.00070 |
| hsa-miR-134-5p_MIMAT0000447 | 156.437 | −1.802 | 3.486 | Down | 0.00258 | 0.02723 |
| hsa-miR-328-5p_MIMAT0026486 | 116.074 | −1.657 | 3.154 | Down | 0.01198 | 0.07091 |
| hsa-let-7f-5p_MIMAT0000067 | 58,027.774 | −1.445 | 2.723 | Down | 0.00046 | 0.01131 |
| hsa-miR-320a_MIMAT0000510 | 4133.844 | −1.432 | 2.698 | Down | 0.00185 | 0.02723 |
| CM000668.1_35903_novel | 361.953 | −1.362 | 2.570 | Down | 0.01030 | 0.07084 |
| hsa-miR-181b-5p_MIMAT0000257 | 1130.724 | −1.259 | 2.393 | Down | 0.00413 | 0.03818 |
| hsa-miR-181b-5p_MIMAT0000257 | 1155.505 | −1.232 | 2.349 | Down | 0.00159 | 0.02723 |
| hsa-miR-409-5p_MIMAT0001638 | 3315.486 | −1.158 | 2.232 | Down | 0.00948 | 0.07019 |
| hsa-miR-744-5p_MIMAT0004945 | 1996.177 | −0.973 | 1.963 | Down | 0.01053 | 0.07084 |
| hsa-miR-181a-5p_MIMAT0000256 | 34,050.461 | −0.656 | 1.575 | Down | 0.01379 | 0.07560 |
| hsa-miR-181a-5p_MIMAT0000256 | 34,050.461 | −0.656 | 1.575 | Down | 0.01379 | 0.07560 |
| Patients | miRNAs | AUC | 95% CI | p-Value | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| STEMI | miR-183-5p | 0.509 | 0.285 to 0.731 | 0.947 | 33.3 | 83.3 |
| miR-134-5p | 0.796 | 0.566 to 0.938 | 0.004 ** | 66.7 | 83.3 | |
| miR-15a-5p | 0.796 | 0.566 to 0.938 | 0.003 ** | 100.0 | 58.3 | |
| let-7i-5p | 0.833 | 0.608 to 0.958 | 0.0005 ** | 100.0 | 66.7 | |
| miR-375 | 0.620 | 0.386 to 0.820 | 0.335 | 88.9 | 41.7 | |
| miR-1307-5p | 0.698 | 0.455 to 0.880 | 0.103 | 100.0 | 58.3 | |
| NSTEMI | miR-183-5p | 0.917 | 0.680 to 0.995 | <0.0001 *** | 100.0 | 75.0 |
| miR-134-5p | 0.717 | 0.451 to 0.904 | 0.144 | 60.0 | 83.3 | |
| miR-15a-5p | 0.617 | 0.355 to 0.836 | 0.411 | 100.0 | 50.0 | |
| let-7i-5p | 0.583 | 0.325 to 0.812 | 0.592 | 80.0 | 83.7 | |
| miR-375 | 0.683 | 0.418 to 0.882 | 0.207 | 60.0 | 83.3 | |
| miR-1307-5p | 0.667 | 0.402 to 0.871 | 0.205 | 100.0 | 50.0 |
| Patients | miRNAs | AUC | 95% CI | p-Value | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| STEMI | miR-183-5p | 0.546 | 0.318 to 0.761 | 0.756 | 55.6 | 75.0 |
| miR-134-5p | 0.769 | 0.535 to 0.922 | 0.027 * | 55.6 | 100.0 | |
| miR-15a-5p | 0.907 | 0.700 to 0.989 | <0.0001 *** | 100.0 | 66.7 | |
| let-7i-5p | 0.929 | 0.722 to 0.995 | <0.0001 *** | 100.0 | 81.8 | |
| NSTEMI | miR-183-5p | 0.917 | 0.680 to 0.995 | <0.0001 *** | 100.0 | 83.3 |
| miR-134-5p | 0.633 | 0.370 to 0.848 | 0.486 | 40.0 | 100.0 | |
| miR-15a-5p | 0.717 | 0.451 to 0.904 | 0.100 | 100.0 | 50.0 | |
| let-7i-5p | 0.618 | 0.348 to 0.843 | 0.525 | 60.0 | 81.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, K.-L.; Mahmood Zuhdi, A.S.; Wan Ahmad, W.A.; Vanhoutte, P.M.; De Magalhaes, J.P.; Mustafa, M.R.; Wong, P.-F. Circulating MicroRNAs in Young Patients with Acute Coronary Syndrome. Int. J. Mol. Sci. 2018, 19, 1467. https://doi.org/10.3390/ijms19051467
Tong K-L, Mahmood Zuhdi AS, Wan Ahmad WA, Vanhoutte PM, De Magalhaes JP, Mustafa MR, Wong P-F. Circulating MicroRNAs in Young Patients with Acute Coronary Syndrome. International Journal of Molecular Sciences. 2018; 19(5):1467. https://doi.org/10.3390/ijms19051467
Chicago/Turabian StyleTong, Kind-Leng, Ahmad Syadi Mahmood Zuhdi, Wan Azman Wan Ahmad, Paul M. Vanhoutte, Joao Pedro De Magalhaes, Mohd Rais Mustafa, and Pooi-Fong Wong. 2018. "Circulating MicroRNAs in Young Patients with Acute Coronary Syndrome" International Journal of Molecular Sciences 19, no. 5: 1467. https://doi.org/10.3390/ijms19051467





