Screening of Natural Product Derivatives Identifies Two Structurally Related Flavonoids as Potent Quorum Sensing Inhibitors against Gram-Negative Bacteria
Abstract
:1. Introduction
2. Results
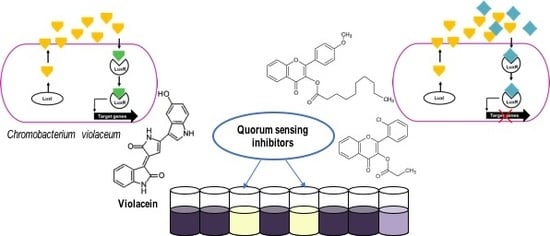
2.1. Primary Screening for Quorum Sensing Inhibitors (QSIs) Using Two Strains of C. violaceum as Reporter Bacteria and Classification of the Compounds
2.2. Highly Active QSIs
2.3. Selection and Characterization of the Leads
2.4. Impact of the Leads on Biofilm Maturation and Architecture in Other Gram-Negative Species
2.5. Impact of the Leads on Swarming and Swimming Motility
2.6. Impact of the Leads on Growth of P. aeruginosa and E. coli
2.7. Cytotoxicity of the Leads
3. Materials and Methods
3.1. Compound Library
3.2. Bacterial Strains, Media and Culture Conditions
3.3. Screening for Quorum Sensing Inhibitors (QSIs)
3.3.1. Violacein Extraction
3.3.2. Resazurin Staining Assay
3.4. Crystal Violet Staining Assay
3.5. Transmitted Light Microscopy
3.6. Swarming and Swimming Motility Assays
3.7. Viable Plate Counts
3.8. Cytotoxicity Testing
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| QS | Quorum sensing |
| QSI | Quorum sensing inhibitor |
| AHL | N-acyl-l-homoserine lactone |
| AI | Autoinducer |
| HSL | Homoserine lactone |
| Quorum quencher | |
| LBY | Luria–Bertani broth supplemented with yeast extract |
| QUE | Quercetin |
References
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- González, J.E.; Keshavan, N.D. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 2006, 70, 859–875. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- McDougald, D.; Rice, S.A.; Kjelleberg, S. Bacterial quorum sensing and interference by naturally occurring biomimics. Anal. Bioanal. Chem. 2007, 387, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Winans, S.C.; Greenberg, E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 1996, 50, 727–751. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T.R.; Gillis, R.; Marx, S.; Brown, C.; Iglewski, B.H. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: Their role and expression patterns. Appl. Environ. Microbiol. 2001, 67, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; O’Neill, J.C.; Blackwell, H.E. Expanding dialogues: From natural autoinducers to non-natural analogues that modulate quorum sensing in gram-negative bacteria. Chem. Soc. Rev. 2008, 37, 1432–1447. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Givskov, M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V. Novel approaches to bacterial infection therapy by interfering with bacteria-to-bacteria signaling. Expert Rev. Anti-Infect. Ther. 2007, 5, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, F.; Fratianni, F.; Coppola, R. Quorum sensing and phytochemicals. Int. J. Mol. Sci. 2013, 14, 12607–12619. [Google Scholar] [CrossRef] [PubMed]
- Givskov, M.; de Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Manefield, M.; de Nys, R.; Kumar, N.; Read, R.; Givskov, M.; Steinberg, P.; Kjelleberg, S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999, 145 (Pt 2), 283–291. [Google Scholar] [CrossRef] [PubMed]
- Manefield, M.; Rasmussen, T.B.; Henzter, M.; Andersen, J.B.; Steinberg, P.; Kjelleberg, S.; Givskov, M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 2002, 148, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Vermote, A.; Brackman, G.; Risseeuw, M.D.; Coenye, T.; van Calenbergh, S. Novel hamamelitannin analogues for the treatment of biofilm related MRSA infections—A scaffold hopping approach. Eur. J. Med. Chem. 2017, 127, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Celen, S.; Hillaert, U.; van Calenbergh, S.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS ONE 2011, 6, e16084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsen, T.H.; Bragason, S.K.; Phipps, R.K.; Christensen, L.D.; van Gennip, M.; Alhede, M.; Skindersoe, M.; Larsen, T.O.; Høiby, N.; Bjarnsholt, T.; et al. Food as a source for quorum sensing inhibitors: Iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.H.; van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annapoorani, A.; Umamageswaran, V.; Parameswari, R.; Pandian, S.K.; Ravi, A.V. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR receptor proteins of Pseudomonas aeruginosa. J. Comput. Aided Mol. Des. 2012, 26, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Skogman, M.E.; Kanerva, S.; Manner, S.; Vuorela, P.M.; Fallarero, A. Flavones as quorum sensing inhibitors identified by a newly optimized screening platform using Chromobacterium violaceum as reporter bacteria. Molecules 2016, 21, 1211. [Google Scholar] [CrossRef] [PubMed]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143 (Pt 12), 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Burt, S.A.; Ojo-Fakunle, V.T.; Woertman, J.; Veldhuizen, E.J. The natural antimicrobial carvacrol inhibits quorum sensing in Chromobacterium violaceum and reduces bacterial biofilm formation at sub-lethal concentrations. PLoS ONE 2014, 9, e93414. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.H.; Rukayadi, Y.; Hwang, J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Deng, Y.; Wang, H.; Liu, W.; Zhuang, X.; Chu, W. Tea polyphenols as an antivirulence compound disrupt quorum-sensing regulated pathogenicity of Pseudomonas aeruginosa. Sci. Rep. 2015, 5, 16158. [Google Scholar] [CrossRef] [PubMed]
- Vattem, D.A.; Mihalik, K.; Crixell, S.H.; McLean, R.J. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia 2007, 78, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Taganna, J.C.; Quanico, J.P.; Perono, R.M.; Amor, E.C.; Rivera, W.L. Tannin-rich fraction from Terminalia catappa inhibits quorum sensing (QS) in Chromobacterium violaceum and the QS-controlled biofilm maturation and LasA staphylolytic activity in Pseudomonas aeruginosa. J. Ethnopharmacol. 2011, 134, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, S.; Nithya, C.; Pandian, S.K. 2-furaldehyde diethyl acetal from tender coconut water (Cocos nucifera) attenuates biofilm formation and quorum sensing-mediated virulence of Chromobacterium violaceum and Pseudomonas aeruginosa. Biofouling 2015, 31, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Manner, S.; Skogman, M.; Goeres, D.; Vuorela, P.; Fallarero, A. Systematic exploration of natural and synthetic flavonoids for the inhibition of Staphylococcus aureus biofilms. Int. J. Mol. Sci. 2013, 14, 19434–19451. [Google Scholar] [CrossRef] [PubMed]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002, 96, 67–202. [Google Scholar] [CrossRef]
- Cushnie, T.P.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Vasavi, H.S.; Arun, A.B.; Rekha, P.D. Anti-quorum sensing activity of Psidium guajava L. flavonoids against Chromobacterium violaceum and Pseudomonas aeruginosa PAO1. Microbiol. Immunol. 2014, 58, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Rekha, P.D.; Vasavi, H.S.; Vipin, C.; Saptami, K.; Arun, A.B. A medicinal herb Cassia alata attenuates quorum sensing in Chromobacterium violaceum and Pseudomonas aeruginosa. Lett. Appl. Microbiol. 2017, 64, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin influences quorum sensing in food borne bacteria: In-vitro and in-silico evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef] [PubMed]
- Truchado, P.; Giménez-Bastida, J.A.; Larrosa, M.; Castro-Ibáñez, I.; Espín, J.C.; Tomás-Barberán, F.A.; García-Conesa, M.T.; Allende, A. Inhibition of quorum sensing (QS) in Yersinia enterocolitica by an orange extract rich in glycosylated flavanones. J. Agric. Food Chem. 2012, 60, 8885–8894. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stévigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rajaonson, S.; Diallo, B.; Mol, A.; El Jaziri, M.; Baucher, M. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2010, 76, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Serra, S.; Cristina Abreu, A.; Saavedra, M.J.; Salgado, A.; Simões, M. Evaluation of the effects of selected phytochemicals on quorum sensing inhibition and in vitro cytotoxicity. Biofouling 2014, 30, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Ryu, Y.B.; Curtis-Long, M.J.; Yuk, H.J.; Cho, J.K.; Kim, J.Y.; Kim, K.D.; Lee, W.S.; Park, K.H. Flavanones and rotenoids from the roots of Amorpha fruticosa L. that inhibit bacterial neuraminidase. Food Chem. Toxicol. 2011, 49, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Celen, S.; Baruah, K.; Bossier, P.; van Calenbergh, S.; Nelis, H.J.; Coenye, T. Ai-2 quorum-sensing inhibitors affect the starvation response and reduce virulence in several Vibrio species, most likely by interfering with LuxPQ. Microbiology 2009, 155, 4114–4122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodini, S.F.; Manfredini, S.; Epp, M.; Valentini, S.; Santori, F. Quorum sensing inhibition activity of garlic extract and p-coumaric acid. Lett. Appl. Microbiol. 2009, 49, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Choi, H.; Hwang, I.S.; Kim, A.R.; Woo, E.R.; Lee, D.G. Synergistic antibacterial and antibiofilm effect between (+)-medioresinol and antibiotics in vitro. Appl. Biochem. Biotechnol. 2013, 170, 1934–1941. [Google Scholar] [CrossRef] [PubMed]
- Monte, J.; Abreu, A.C.; Borges, A.; Simões, L.C.; Simões, M. Antimicrobial activity of selected phytochemicals against Escherichia coli and Staphylococcus aureus and their biofilms. Pathogens 2014, 3, 473–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo-Romano, B.; Gollihar, J.; Brown, S.A.; Whiteley, M.; Valenzuela, E.; Kaplan, H.B.; Wood, T.K.; McLean, R.J. Indole inhibition of N-acylated homoserine lactone-mediated quorum signalling is widespread in gram-negative bacteria. Microbiology 2014, 160, 2464–2473. [Google Scholar] [CrossRef] [PubMed]
- More, P.G.; Karale, N.N.; Lawand, A.S.; Rajmane, S.V.; Pawar, S.V.; Patil, R.H. A 4-(o-methoxyphenyl)-2-aminothiazole: An anti-quorum sensing compound. Med. Chem. Res. 2012, 22, 4183–4191. [Google Scholar] [CrossRef]
- Cho, H.S.; Lee, J.H.; Cho, M.H.; Lee, J. Red wines and flavonoids diminish Staphylococcus aureus virulence with anti-biofilm and anti-hemolytic activities. Biofouling 2015, 31, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Regmi, S.C.; Kim, J.A.; Cho, M.H.; Yun, H.; Lee, C.S.; Lee, J. Apple flavonoid phloretin inhibits Escherichia coli O157:H7 biofilm formation and ameliorates colon inflammation in rats. Infect. Immun. 2011, 79, 4819–4827. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.F.; Ren, L.B.; Teng, Y.; Zheng, S.; Yang, X.L.; Guo, X.J.; Wang, X.Y.; Sha, K.H.; Li, N.; Xu, G.Y.; et al. Luteolin decreases the attachment, invasion and cytotoxicity of UPEC in bladder epithelial cells and inhibits UPEC biofilm formation. Food Chem. Toxicol. 2014, 72, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Justo, G.Z.; Durán, M.; Brocchi, M.; Cordi, L.; Tasic, L.; Castro, G.R.; Nakazato, G. Advances in Chromobacterium violaceum and properties of violacein-its main secondary metabolite: A review. Biotechnol. Adv. 2016, 34, 1030–1045. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem BioAssay Database. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/924 (accessed on 28 March 2018).
- Kwan, J.C.; Meickle, T.; Ladwa, D.; Teplitski, M.; Paul, V.; Luesch, H. Lyngbyoic acid, a “tagged” fatty acid from a marine cyanobacterium, disrupts quorum sensing in Pseudomonas aeruginosa. Mol. Biosyst. 2011, 7, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Marques, C.N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, T.; Kato, M.; Fukamachi, K.; Kato, N.; Ikeda, T. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol. Lett. 2008, 279, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.S.; Iglewski, B.H. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003, 6, 56–60. [Google Scholar] [CrossRef]
- Ahmer, B.M. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 2004, 52, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Bjarnsholt, T.; Skindersoe, M.E.; Hentzer, M.; Kristoffersen, P.; Köte, M.; Nielsen, J.; Eberl, L.; Givskov, M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 2005, 187, 1799–1814. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002, 148, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef] [PubMed]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Das, M.C.; Sandhu, P.; Das, N.; Tribedi, P.; De, U.C.; Akhterb, Y.; Bhattacharjee, S. Antibiofilm activity of Parkia javanica against Pseudomonas aeruginosa: A study with fruit extract. RSC Adv. 2017, 7, 5497–5513. [Google Scholar] [CrossRef]
- O’May, C.; Tufenkji, N. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol. 2011, 77, 3061–3067. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Shingaki, R.; Fukui, K. Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol. Lett. 2008, 281, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Caiazza, N.C.; Shanks, R.M.; O’Toole, G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef] [PubMed]
- Manner, S.; Fallarero, A.; Abo Akademi University and University of Helsinki, Finland. Evaluation of the Effects of the Leads in Rhamnolipid Biosynthesis. Personal communication, 2017. [Google Scholar]
- Issac Abraham, S.V.; Palani, A.; Ramaswamy, B.R.; Shunmugiah, K.P.; Arumugam, V.R. Antiquorum sensing and antibiofilm potential of Capparis spinosa. Arch. Med. Res. 2011, 42, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.C.; Grayston, J.T. A sensitive cell line, HL cells, for isolation and propagation of Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 1990, 162, 755–758. [Google Scholar] [CrossRef] [PubMed]






| Compound | Compound ID | ATCC 31532 | CV026 | Class |
|---|---|---|---|---|
| 144 | ST012391 | 94.7 | 97.2 | Nucleoside analogue |
| 339 | ST024776 | 95.6 | 94.7 | N/A, a mixture of two compounds * |
| 341 | ST024784 | 93.4 | 95.2 | Flavonoid (isoflavonoid) |
| 575 | ST045414 | 94.1 | 94.3 | Flavonoid (flavonol) |
| 916 | ST069294 | 101.9 | 95.7 | Flavonoid (flavone) |
| 1698 | ST077117 | 91.3 | 96.1 | Lignan |
| 2117 | ST083092 | 101.8 | 101.9 | Flavonoid (flavone) |
| 2307 | ST088527 | 98.8 | 98.6 | Alkaloid |
| 2896 | ST079962 | 94.5 | 91.2 | Flavonoid (flavone) |
| Compound | Class | Inhibition-% 1 | IC50 (µM) | 95% Confidence Intervals |
|---|---|---|---|---|
| 144 | Nucleoside analogue | 0 ± 5.2 | N.A. | |
| 339 | N/A | 3.7 ± 1.8 | N.A. | |
| 341 | Flavonoid (isoflavonoid) | 26.9 ± 7.7 | N.A. | |
| 575 | Flavonoid (flavonol) | 60.9 ± 4.1 | N.A. | |
| 916 | Flavonoid (flavone) | 35.8 ± 1.1 | N.A. | |
| 1698 | Lignan | 2.6 ± 4.6 | N.A. | |
| 2117 | Flavonoid (flavone) | 99.7 ± 1.0 | 9.6 | 7.9–11.8 |
| 2307 | Alkaloid | 76.1 ± 3.3 | N.A. | |
| 2896 | Flavonoid (flavone) | 83.5 ± 1.4 | 13.9 | 8.5–22.5 |
| Que | Flavonoid (flavonol) | 91.8 ± 2.3 | 3.1 | 2.1–4.5 |
| Strain | 2117 | 2896 |
|---|---|---|
| P. aeruginosa PAO1 | 42.3 ± 12.4 | 11.6 ± 11.1 |
| P. aeruginosa ATCC 9027 | 27.2 ± 6.0 | 22.5 ± 3.2 |
| P. aeruginosa ATCC 15422 | 11.2 ± 1.8 | 28.1 ± 15.1 |
| E. coli K-12 | 51.3 ± 6.9 | 14.3 ± 10.1 |
| Compound | Concentration | HL | RAW 264.7 |
|---|---|---|---|
| 2117 | 100 µM | 11.2 ± 4.9 | 97.9 ± 0.5 |
| 2896 | N.I. | N.I. | |
| 2117 | 40 µM | N.I. | N.I. |
| 2896 | N.I. | N.I. | |
| 2117 | 10 µM | N.I. | N.I. |
| 2896 | N.I. | N.I. | |
| Usnic acid | 200 µM | 90.0 ± 4.7 | 98.5 ± 0.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manner, S.; Fallarero, A. Screening of Natural Product Derivatives Identifies Two Structurally Related Flavonoids as Potent Quorum Sensing Inhibitors against Gram-Negative Bacteria. Int. J. Mol. Sci. 2018, 19, 1346. https://doi.org/10.3390/ijms19051346
Manner S, Fallarero A. Screening of Natural Product Derivatives Identifies Two Structurally Related Flavonoids as Potent Quorum Sensing Inhibitors against Gram-Negative Bacteria. International Journal of Molecular Sciences. 2018; 19(5):1346. https://doi.org/10.3390/ijms19051346
Chicago/Turabian StyleManner, Suvi, and Adyary Fallarero. 2018. "Screening of Natural Product Derivatives Identifies Two Structurally Related Flavonoids as Potent Quorum Sensing Inhibitors against Gram-Negative Bacteria" International Journal of Molecular Sciences 19, no. 5: 1346. https://doi.org/10.3390/ijms19051346