1. Introduction
The monitoring of blood glucose is essential for diabetes therapy. Recently, more attention has been given to continuous glucose monitoring (CGM), in which the glucose concentration of interstitial fluid is measured continuously by a glucose sensor inserted subcutaneously or by non-invasive methods [
1]. CGM together with the combination of sensor augmented insulin infusion technologies is currently the most advanced biomedical engineering in diabetes therapy. Recently, the U.S. Food and Drug Administration approved the first automated insulin delivery device for type 1 diabetes [
2]. Conventional enzyme sensors require electron mediators to transfer electrons to the electrode. By contrast, DET-type enzyme sensors transfer electrons directly from the enzyme to the electrode. The DET principle enables the construction of lower potential sensors and avoids the use of toxic mediators in the system, thus making it attractive for implantable sensors. Glucose sensors employing enzymes that are capable of DET will be ideal for the application for CGM.
FAD-dependent glucose dehydrogenase isolated from
Burkholderia cepacia (FADGDH) [
3] is one of the limited glucose dehydrogenase enzymes capable of DET with an electrode. This enzyme is a hetero-oligomeric enzyme composed of three subunits: a catalytic subunit (α subunit) harboring an FAD cofactor, a small subunit (γ subunit) required for proper folding and secretion of the α subunit, and a cyt
c subunit (β subunit), which harbors three hemes (heme 1, heme 2, and heme 3, from the N-terminal sequence). Cloning and functional expression in
Escherichia coli (
E. coli) was achieved [
4,
5], and protein engineering studies have been conducted to improve its substrate specificity, making this enzyme more suitable for glucose sensing [
6,
7]. Additionally, the development of several glucose sensing devices utilizing the DET capability of this enzyme have been reported [
7,
8,
9,
10]. Recently, the presence of a 3Fe–4S-type iron-sulfur cluster in the α subunit was elucidated [
11]. The 3Fe–4S-type iron-sulfur cluster is the primary electron acceptor of FAD in the catalytic subunit, and it may play a role in the inter-molecular electron transfer from FAD to the multi-heme cyt
c subunit.
To facilitate further engineering of FADGDH as a DET-type glucose sensor component, elucidation of the electron transfer pathway within this enzyme is necessary. Previous studies have revealed that when FADGDH is immobilized onto an electrode without electron mediators, the current response for glucose is drastically lowered in the absence of the β subunit [
8], thus demonstrating that DET occurs via the β subunit. The spectrum unique to cyt
c changed to its reduced form with the addition of glucose to the FADGDH solution [
12]. From these results it is obvious that the electrons generated by the catalytic reaction of glucose oxidation are transferred from the α subunit to the β subunit. Combining these facts with the recent finding of the presence of the iron-sulfur cluster in the α subunit [
11], it could be presumed that the electrons generated by the catalytic reactions are transferred from FAD through the iron-sulfur cluster to the β subunit and then to the electrode in the DET reaction. Although electrons are likely transferred to the electrode from one of the three covalently bound hemes of the β subunit, the detailed electron pathway within the β subunit remains unknown.
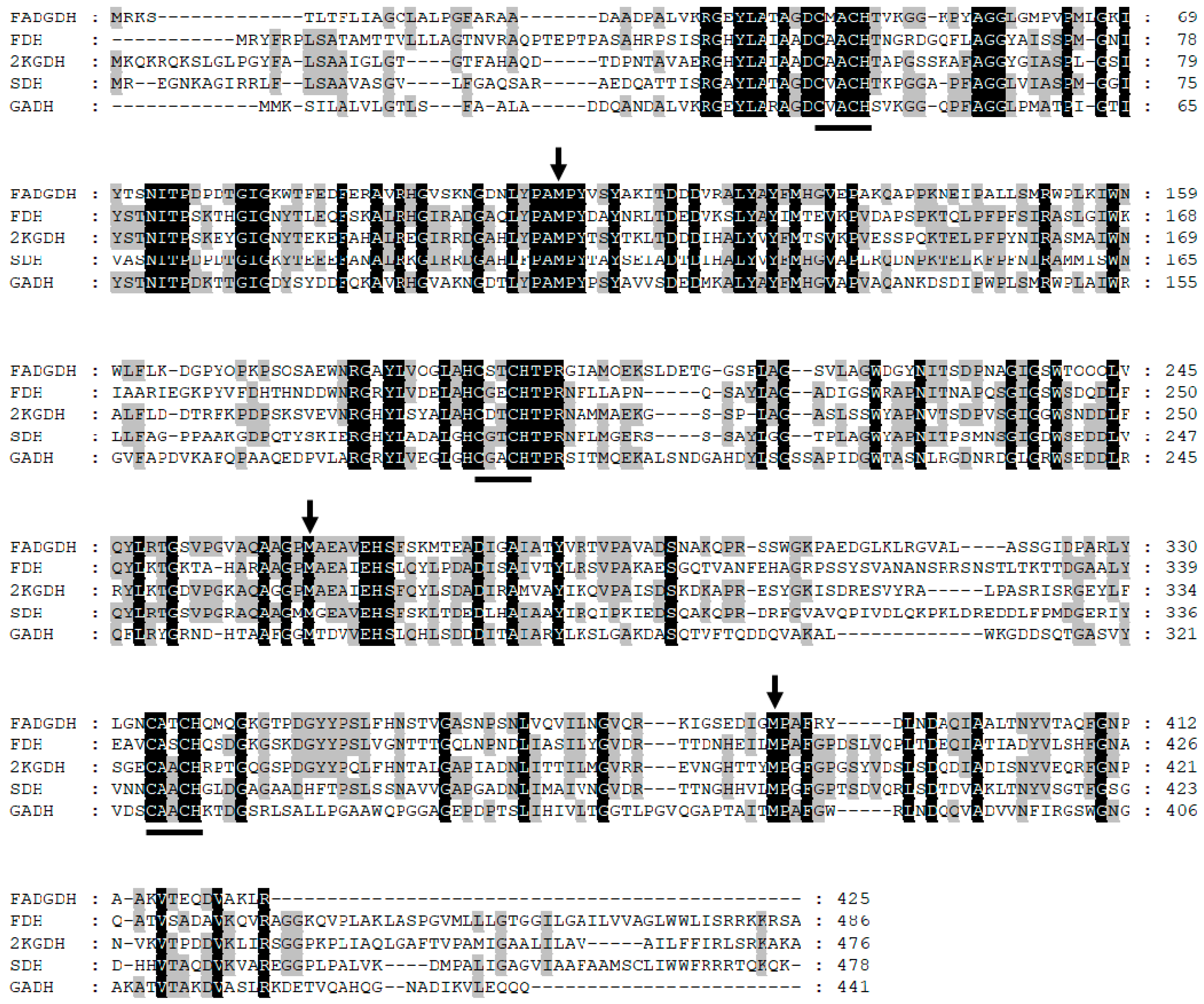
Elucidation of the inter/intra-electron transfer pathway in the β subunit will reveal the entire pathway from glucose oxidation to the external electron acceptor. To achieve this goal, a mutagenesis study of the residues proximal to the hemes was conducted. By comparing and analyzing the primary structures of the β subunit and the cyt
c subunits of the reported hetero-oligomeric flavocytochrome dehydrogenases using the Clustal Omega software (Available online:
https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 17 Feb 2018) [
13], the sixth ligand for each of the three heme irons was predicted (
Figure 1). Consequently, Met109, Met263, and Met386 were substituted to His. The catalytic activities of the WT and mutant enzymes were compared by investigating their dye-mediated dehydrogenase activities and their DET abilities toward the electrode. Based on the obtained results, two different electron transfer pathways were proposed depending on the electron acceptor.
3. Discussion
In this study, we aimed to elucidate the inter/intra-electron transfer pathway in the β subunit of FADGDH. To the best of our knowledge, no 3D structure of FAD dependent dehydrogenase composed of cyt
c subunit was elucidated. In addition, no homologous structure to the β subunit of FADGDH was reported. However, the analysis of FADGDH has revealed that the dissociation of β subunit occurs after heat treatment resulting in the decrease of enzymatic activity showing that the β subunit plays an important role in the electron transfer between α subunit and the artificial electron mediator [
12]. Cross-linking of the subunits of FADGDH resulted in high thermal stability without loss of catalytic activity supporting this hypothesis [
19]. Therefore, the formation of quaternary structure of FADGDH is inevitable to show its characteristic electron transfer via cyt
c subunit.
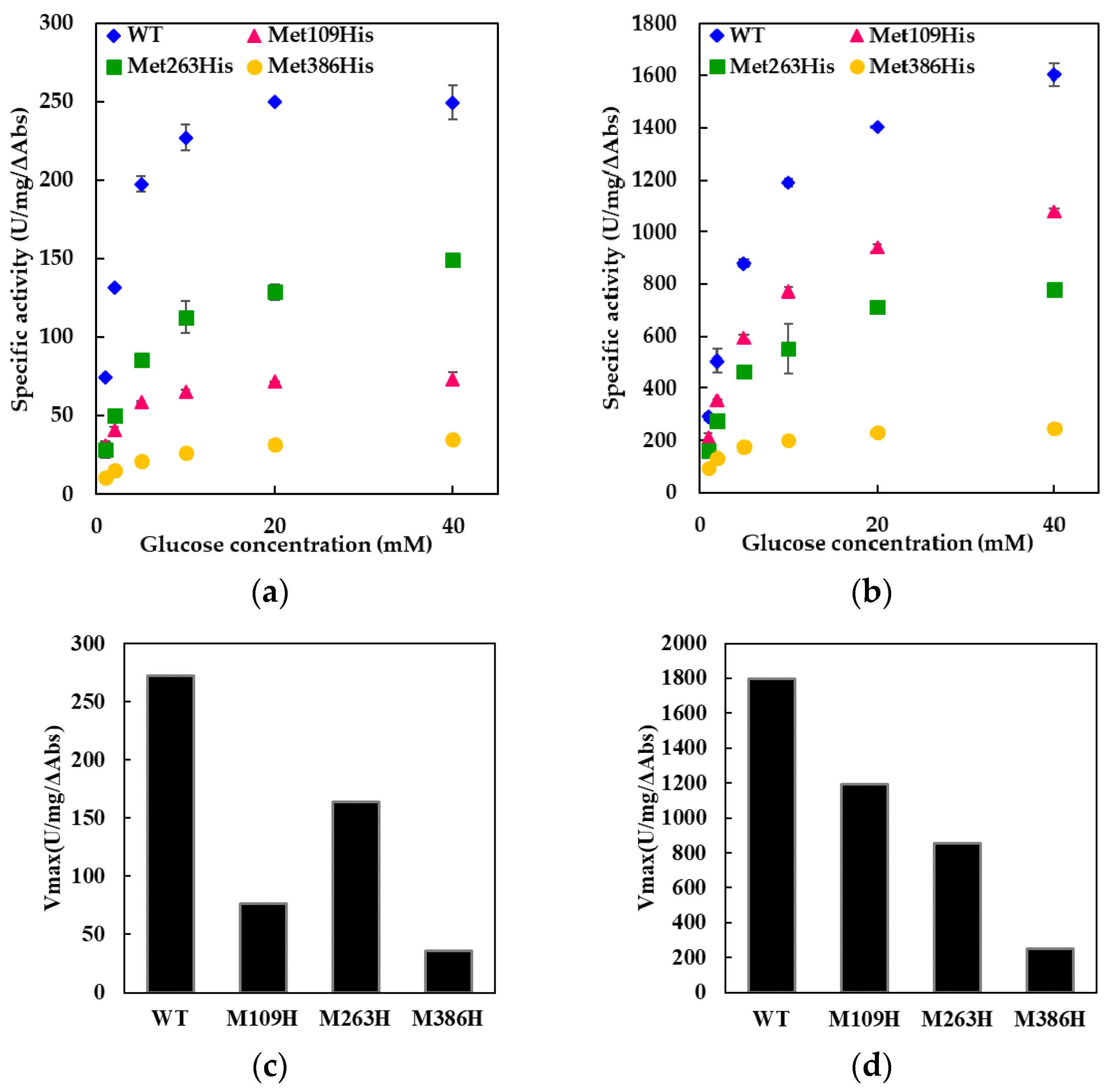
Throughout our experiments using FADGDH with electron transfer subunit, no precipitation was observed. The mutated electron transfer subunits were properly folded, thereby forming active quaternary structure with catalytic subunit. This was confirmed by the observation of the electron transfer reaction by the oxidation of glucose at the catalytic subunit resulting in reduction of electron acceptor, especially 2-hexaamineruthenium(III) chloride. In the activity assay using 2-hexaamineruthenium(III) chloride as an electron acceptor, the electrons can only be transferred from the β subunit, but not from catalytic subunit alone (
Table S1; the comparison of dye-mediated dehydrogenase activities of catalytic subunit (γα), catalytic subunit with wild-type electron transfer subunit (β), and of catalytic subunit with mutated electron transfer subunits (β)). With the catalytic subunit alone, the dehydrogenase activities using 2-hexaamineruthenium(III) chloride was almost negligible compared with those using PMS-DCPIP. In contrary, the dehydrogenase activity of the catalytic subunit with the electron transfer subunit using 2-hexaamineruthenium(III) chloride was about 18% for wild type, 7.7% for Met109His mutant, 18% for Met263His mutant, and 14% for Met386His mutant compared with the assay using PMS-DCPIP. Thus, it is obvious that 2-hexaamineruthenium(III) chloride-dependent dye mediated dehydrogenase activity can only be seen when the catalytic subunit is forming a complex with the β subunit. We believe that no enzyme quaternary structure will be formed if the electron transfer subunit is not correctly folded. Therefore, these results simultaneously suggested that all mutant electron transfer subunits were folded properly to form quaternary structure, and thereby showed 2-hexaamineruthenium(III) chloride dependent dehydrogenase activity.
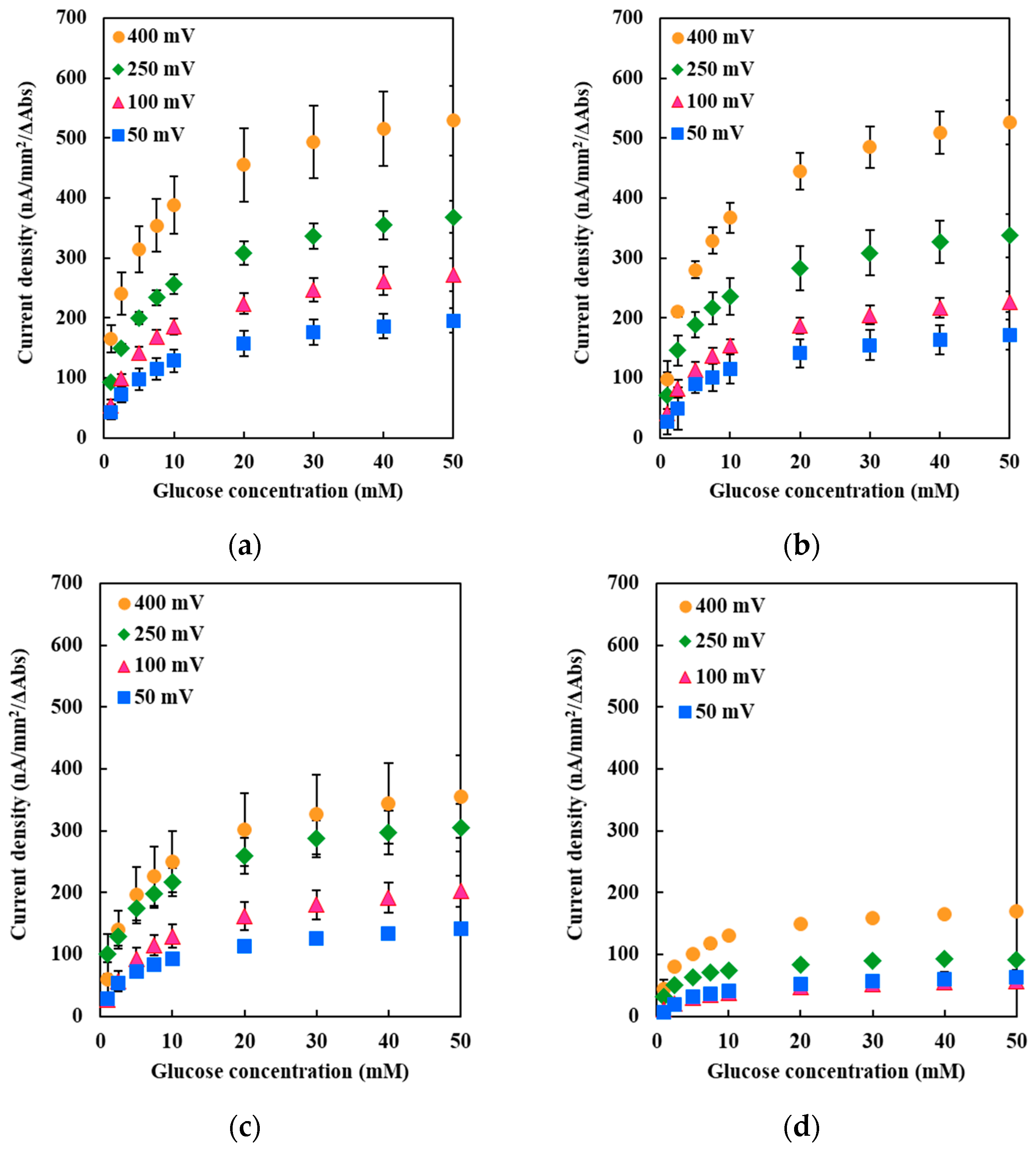
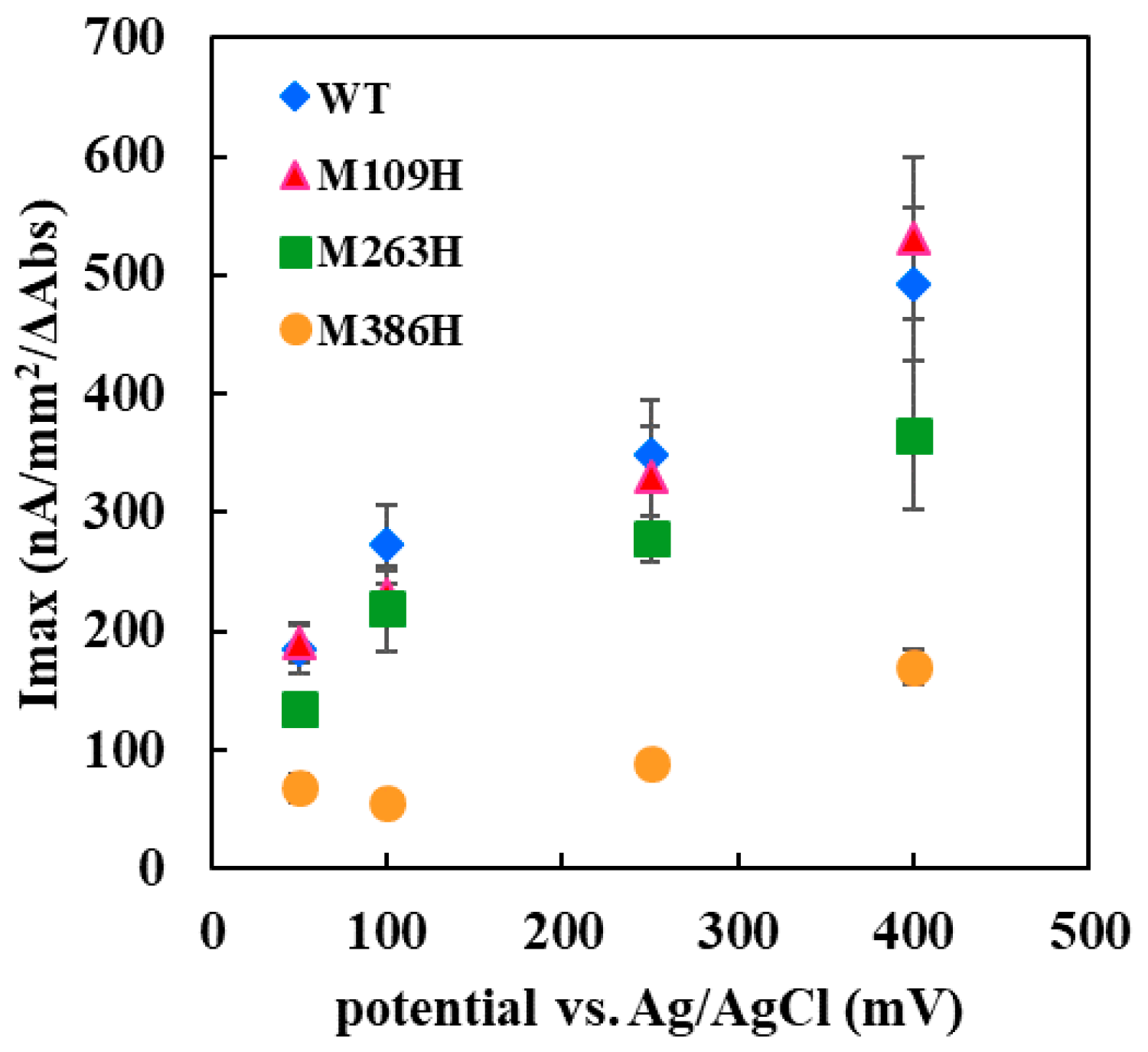
The Met109His mutation on the sixth ligand of heme 1 of FADGDH resulted in a change in the mediator dependent dye-mediated dehydrogenase, but did not affect the DET ability toward the electrode of this enzyme. The Met263His mutation on the sixth ligand of heme 2 of FADGDH did not result in a significant change in the mediator dependent dye-mediated dehydrogenase activity, whereas the Met263His mutation affected the DET ability, especially the applied potential dependency. Considering that the reaction conditions of enzyme with electrodes themselves are artificial, and would vary depending on the way of immobilization and the electrode materials, this enzyme complex originally may transfer electron via heme 1, and the role of heme 2 to transfer electron to electrode is observed only at the artificial conditions
The Met386His mutation on the sixth ligand of heme 3 of FADGDH resulted not only in a drastic decrease in the dye-mediated dehydrogenase activity regardless of the electron acceptors, but also the DET ability. The β subunit is essential to show 2-hexaammineruthenium(III) chloride-mediated dehydrogenase activity. Since this activity was obviously high compared with the FADGDH without β subunit (
Table S1), it is evident that the Met386His mutant accepts electron from catalytic subunit by retaining its quaternary structure, and the electrons are transferred to external electron acceptor via β subunit.
The dye-mediated dehydrogenase activity was affected by the mutation in all three constructed mutants, it could be presumed that all hemes in the β subunit are involved in the electron transfer in the β subunit. Since the catalytic reaction occurs at the α subunit, the electrons are passed on to the β subunit and then to the external electron acceptors. The information allowed us to give an assumption that each heme of the FADGDH β subunit should have different roles; one responsible for the electron acceptor from the catalytic subunit, another one responsible for internal electron transfer and the last one responsible for the electron transfer to the external electron acceptor.
Only the mutation in the sixth ligand of heme 1 affected the response toward the two different electron acceptors used for the dye-mediated dehydrogenase assays. As the mutation in heme responsible for the electron transfer to the external electron acceptors might cause such an alteration, heme 1 was presumed to be mainly responsible to donate electron to the external electron acceptors in solution. Surprisingly, the DET ability observed for the electrode immobilized with the Met109His mutant was not affected by the mutation. Thus, the assumption that heme 1 is not involved in the DET could be made. If the mutation at the electron accepting heme resulted in a negative impact on the electron transfer from catalytic subunit, the effect should appear both in dye-mediated dehydrogenase activity and DET ability. The effect of the mutation in the sixth ligand of heme 3 resulted in a drastic decrease in the dye-mediated dehydrogenase activity and also the DET ability. Since Met109His mutation had a negative effect on dye-mediated dehydrogenase activity, but did not affect the DET ability, heme 1 is not the electron accepting heme. The Met263His mutation also had negative impact on both the dye-mediated dehydrogenase activity and the DET ability but the Met386His mutation had more drastic effect. Therefore, it was presumed that heme 3 may be responsible for accepting electrons from the α subunit. Since the presence of 3Fe–4S-type iron-sulfur cluster in the α subunit is reported in previous studies [
11], the electrons may be transferred via the iron-sulfur cluster. This mutation may have caused a shift in the redox potential of heme 3, thus reduced the internal electron transfer efficiency. Although the mutation in the sixth ligand of heme 2 showed a slight decrease in the dye-mediated dehydrogenase activity, the impact was the same toward both electron acceptors investigated in this study, unlike for Met109His. Therefore, heme 2 does not seem to be the main heme responsible for external electron transfer in solution but is only responsible for internal electron transfer. However, Met263His affected the DET ability, especially the applied potential dependency. From these observations, heme 2 could be presumed to be the heme responsible for DET to the electrode.
Considering these observations, the following two schemes for the electron transfer pathway are proposed, which are dependent on the availability of electron acceptors, soluble electron acceptors (
Figure 5a) or an electrode for direct electron transfer (
Figure 5b). First pathway is the electron transfer to electron acceptors dissolved in solution. In this pathway, the electrons are first passed from the α subunit via the iron sulfur cluster to heme 3, then to heme 2, and finally to heme 1. Although we cannot rule out the possibility that heme 2 may transfer electron directly to the external electron acceptor, heme 1 is the main electron-donating heme for the external electron acceptor in this pathway. The second pathway is the electron transfer pathway for DET electrocatalysis. The electron from the α subunit is also transferred through the iron-sulfur cluster to heme 3 in this pathway and then to heme 2. Heme 1 may accept an electron from heme 2. but it is not involved in DET with the electrode, and heme 2 is the heme responsible for electron transfer to the electrode.
The electron transfer pathway for DET-type bioelectrocatalysis for the cyt
c subunit of fructose dehydrogenase, which is also a hetero-oligomeric flavocytochrome dehydrogenase, has been reported recently [
20,
21,
22]. Through these studies, the authors concluded that the heme 2 is responsible for DET, without the contribution of heme 1. These results are in agreement with the results obtained in this study for FADGDH. Although the study of fructose dehydrogenase did not refer to the role of heme 1, our results indicated that heme 1 in fructose dehydrogenase might be involved in the electron transfer pathway in solution.
The present findings will provide valuable insights into understanding the DET mechanism of FADGDH, which is necessary for the further improvement of the performance and its application toward CGM systems and other DET-based glucose monitoring.
4. Materials and Methods
4.1. Chemicals
Phenazine methosulfate (PMS), D-glucose, and 2,6-dichlorophenolindophenol (DCPIP) were purchased from the Kanto Chemical Co., Ltd. (Tokyo, Japan). Ketjenblack EC600JT was purchased from the Lion Specialty Chemicals Co., Ltd. (Tokyo, Japan). Glutaraldehyde solution (25%, w/v) was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Nafion perfluorinated resin solution, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) and 2-hexaammineruthenium(III) chloride were purchased from Sigma-Aldrich Japan G. K. (Tokyo, Japan). All other chemicals were of reagent grade.
4.2. Site-Directed Mutagenesis
Site-directed mutagenesis was carried out by overlap PCR using pTrcγαATGβ [
5], the expression vector for the FADGDH γαβ complex, as a template. The resulting mutated PCR fragments and pTrcγαATGβ were cut by the restriction enzymes
EcoRI and
NotI, followed by ligation. All mutations were confirmed by nucleotide sequencing.
4.3. Preparation of FADGDH WT, Met109His, Met263His and Met386His
Each of the WT and mutated pTrcγαATGβ expression vectors was co-transformed into
E. coli BL21(DE3) with the pBBJMccm [
5] vector, which encodes the
E. coli ccmABCDEFGH genes. The co-transformed
E. coli were grown aerobically at 37 °C for 30 h in ZYP-5052 auto-induction medium (0.5% glycerol, 0.05% glucose, 0.2% lactose, 50 mM (NH
4)PO
4, 50 mM Na
2HPO
4, and 1 mM MgSO
4) [
23] containing 100 μg/mL ampicillin and 50 μg/mL kanamycin. The harvested cells were washed with 0.85% (
w/
v) NaCl, resuspended in 10 mM potassium phosphate buffer (PPB) at a pH of 7.0 and lysed by French pressing. Lysates were centrifuged at 9000×
g at 4 °C for 15 min, and the obtained supernatants were centrifuged again at 104,000×
g at 4 °C for 1 h. The pellets were suspended in 1 mL of 10 mM PPB (pH 7.0) containing 1.5% (
w/
v) sodium cholate and 0.1 M KCl per 0.5 g of pellet and solubilized at 1600 rpm for 1 h at 4 °C. The solubilized samples were centrifuged at 104,000×
g at 4 °C, and crude extracts were obtained by dialyzing against 10 mM PPB at a pH of 7.0.
FADGDH was purified by hydrophobic interaction chromatography. The crude extracts were applied onto a 5 mL Hitrap Octyl FF Column (GE Healthcare Japan Co., Tokyo, Japan) equilibrated with 10 mM PPB (pH 7.0) and eluted using six column volumes of 10 mM PPB (pH 7.0) containing 0.4% (w/v) sodium cholate, followed by three column volumes of 10 mM PPB (pH 7.0) containing 1.0% (w/v) sodium cholate. The active fractions with absorbance at 410 nm were pooled and dialyzed against 10 mM PPB (pH 7.0).
4.4. Enzyme Activity Assay
The specific activities of FADGDH (WT or mutants) toward various concentrations of glucose were evaluated using two different mediators as the primary external electron acceptors. The electron mediators used were PMS and 2-hexaammineruthenium(III) chloride.
The activity assay using PMS as the electron mediator was carried out using color to indicate the second electron acceptor, DCPIP, by monitoring the decrease in the absorbance of DCPIP at 600 nm as described previously [
11], except the assay buffer contained 0.2% Triton X-100.
The activity assay using the 2-hexaammineruthenium(III) chloride as the electron mediator was carried out in 10 mM PPB (pH 7.0) containing 0.2% Triton X-100 at room temperature with the presence of 1 mM MTT as a color indicator of the second electron acceptor, 2% 2-hexaammineruthenium(III) chloride with various concentrations of glucose. The increase in the absorbance of reduced MTT at 565 nm was recorded.
In both assays one unit (U) of GDH activity was defined as the amount that oxidized 1 μmol of glucose per minute under the standard assay conditions using the millimolar absorption coefficient of 16.3 mM/cm for DCPIP and 20 mM/cm for MTT. In both assays, the protein concentration was measured using a Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA) kit to calculate the specific activity (U/mg).
To better evaluate the effects of mutations on the function of the β subunit, the results from the activity assay was normalized via the absorbance at the soret band (410 nm) of each enzyme (WT and mutants). The specific activity is indicated as U/mg/ΔAbs.
4.5. Electrode Preparation
The purified FADGDH (WT or mutants) was diluted to 0.2 mg/mL by 100 mM PPB (pH 7.0). This solution, 5% (w/v) Nafion and Ketjenblack EC600JT dispersed in 0.8% Triton X-100 were mixed together in a ratio of 8:1:1, respectively. One microliter of this mixture was applied onto a glassy carbon electrode (7 mm2) five times to apply 5 µL in total. The electrode was air-dried at 4 °C degrees for 1 h and was then placed in a sealed container filled with vapor from 25% (w/v) glutaraldehyde for 90 min of cross-linking. The electrode was stored at 25 °C until use.
4.6. Electrochemical Measurements
Chronoamperometry was carried out with the constructed enzyme electrodes in a three-electrode system: FADGDH (WT or mutants), as the immobilized working electrode; Ag/AgCl (3 M NaCl), as the reference electrode (model RE-1, BAS, Tokyo, Japan); and Pt wire, as the counter electrode. All three electrodes were placed in a water jacket cell filled with 10 mL of reaction buffer and 100 mM PPB (pH 7.0), and they were kept at 37 °C. The reaction buffer was continuously stirred at 250 rpm during the measurement. Potentials of +50, +100, +250, and +400 mV vs. Ag/AgCl were applied, and the current responses with the presence of various glucose concentrations (1, 2.5, 5, 7.5, 10, 20, 30 and 40 mM) in the reaction buffer were measured. The glucose concentration was increased by adding the glucose solution to the reaction buffer. The current density was calculated from the area of the working electrode (7 mm2).
The results for electrochemical measurements were also normalized via the absorbance at the soret band of each enzyme as described previously in
Section 4.4. The current density is indicated as nA/mm
2/ΔAbs.