Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx mori
Abstract
:1. Introduction
2. Results and Discussion
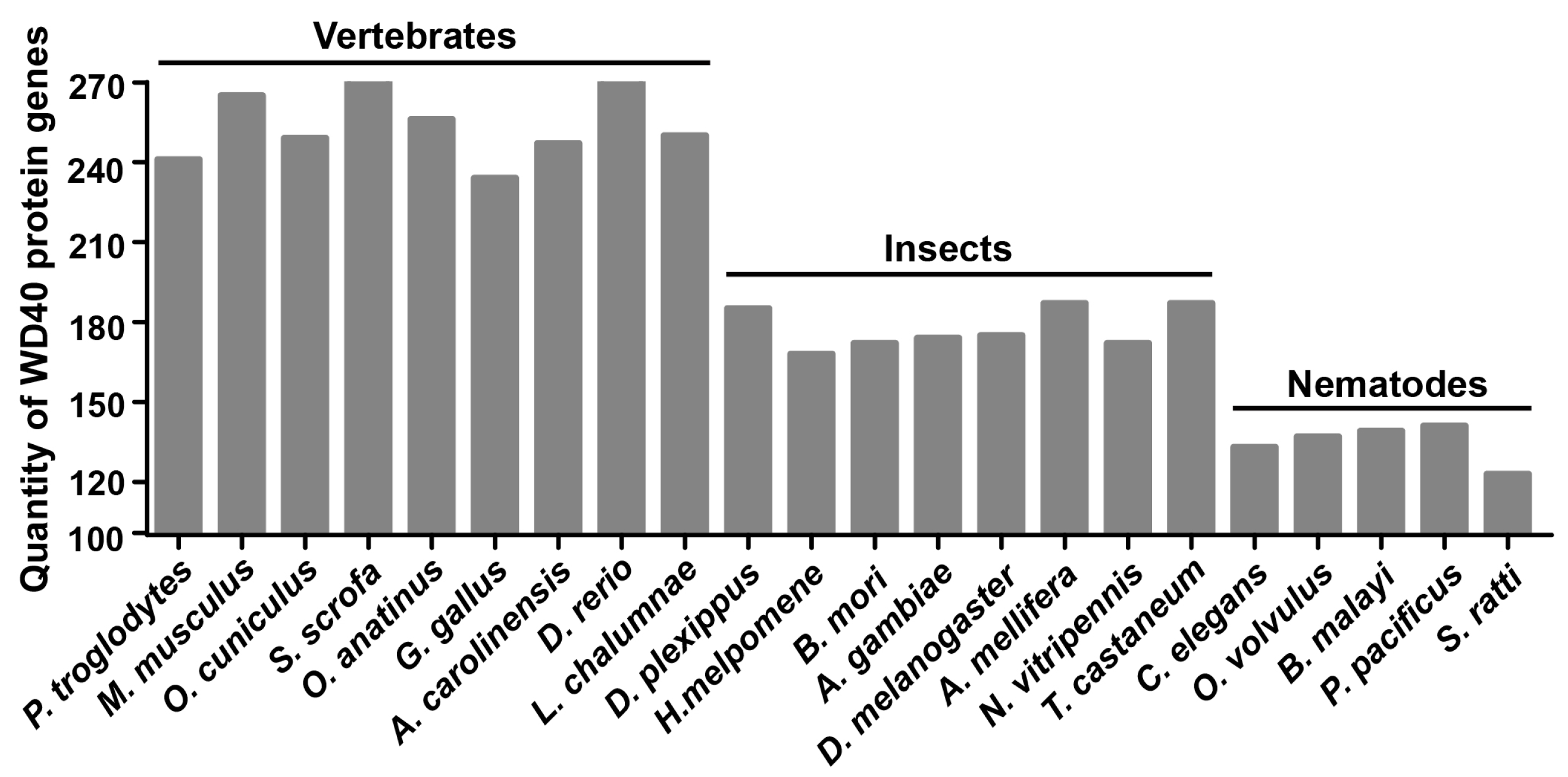
2.1. Identification of WD40 Proteins
2.2. Chromosomal Distribution and Gene Structure of the BmWD40 Family
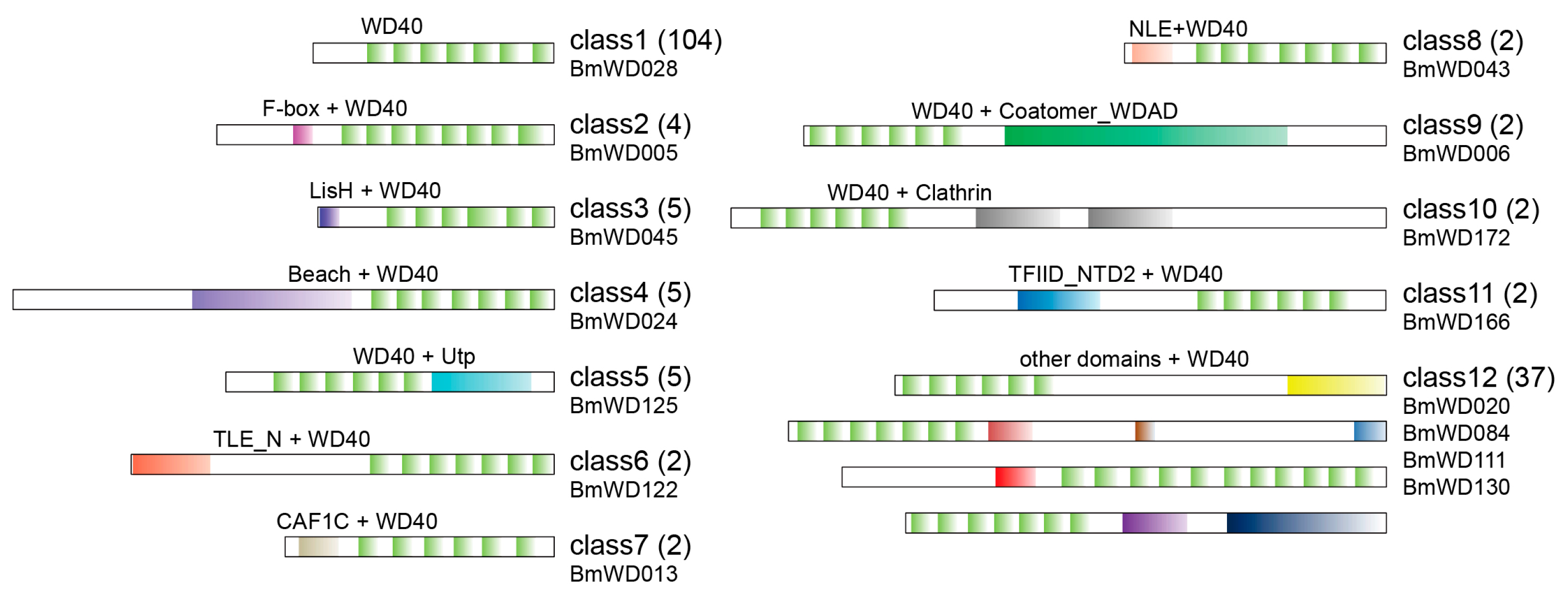
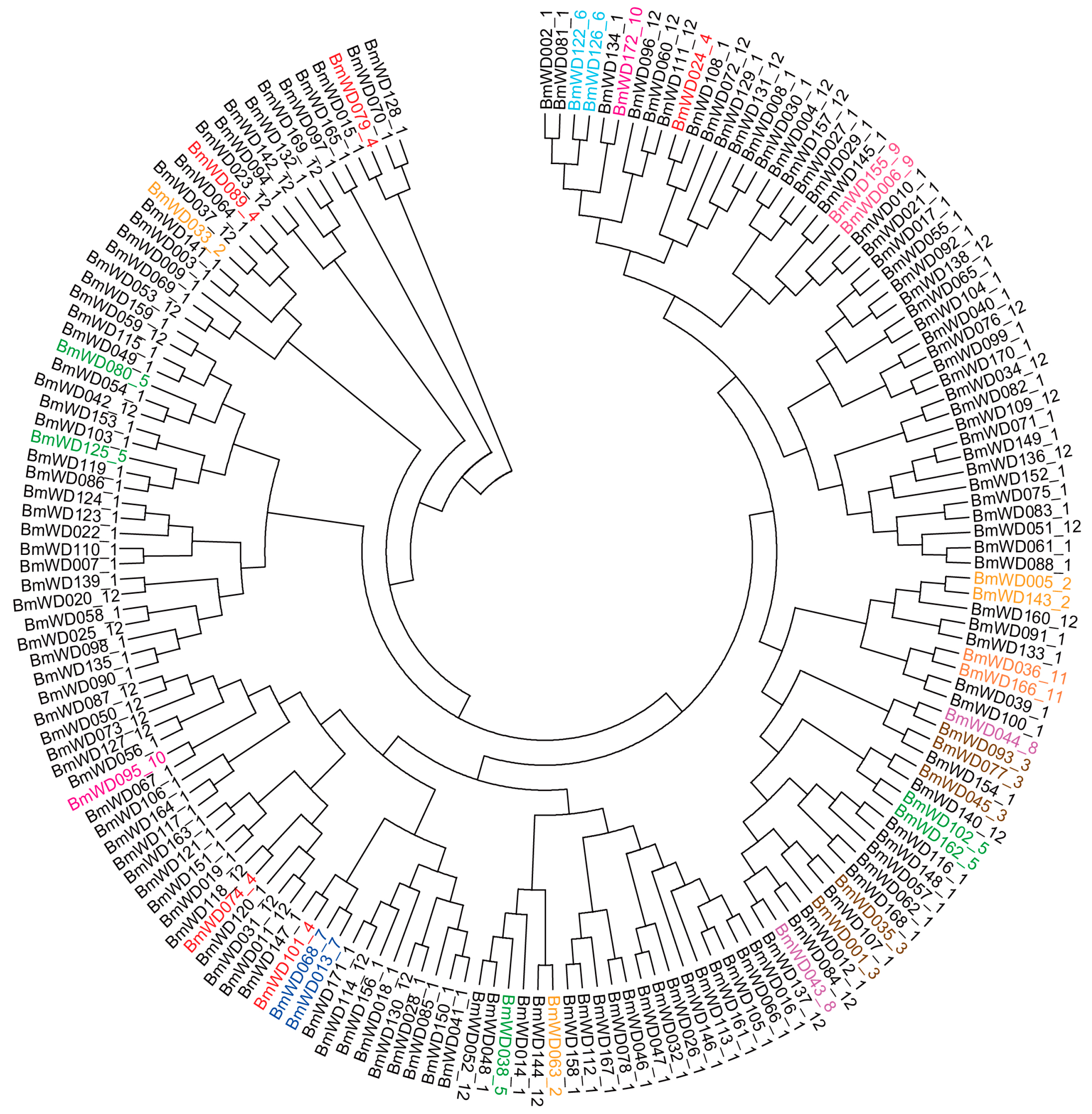
2.3. Phylogenetic Classification and Domain Architectures of BmWD40s
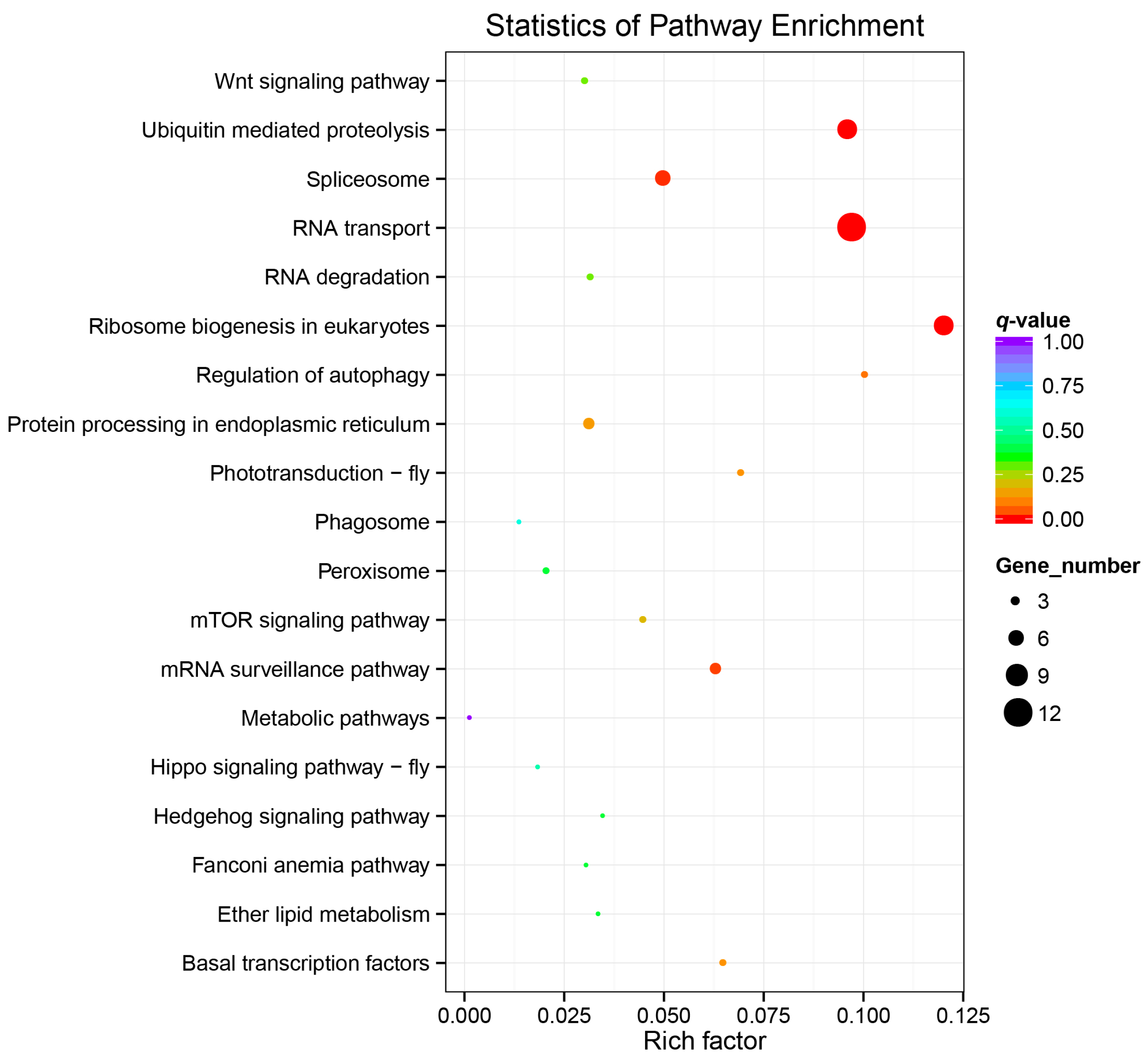
2.4. Gene Ontology and KEGG Analysis of the BmWD40s
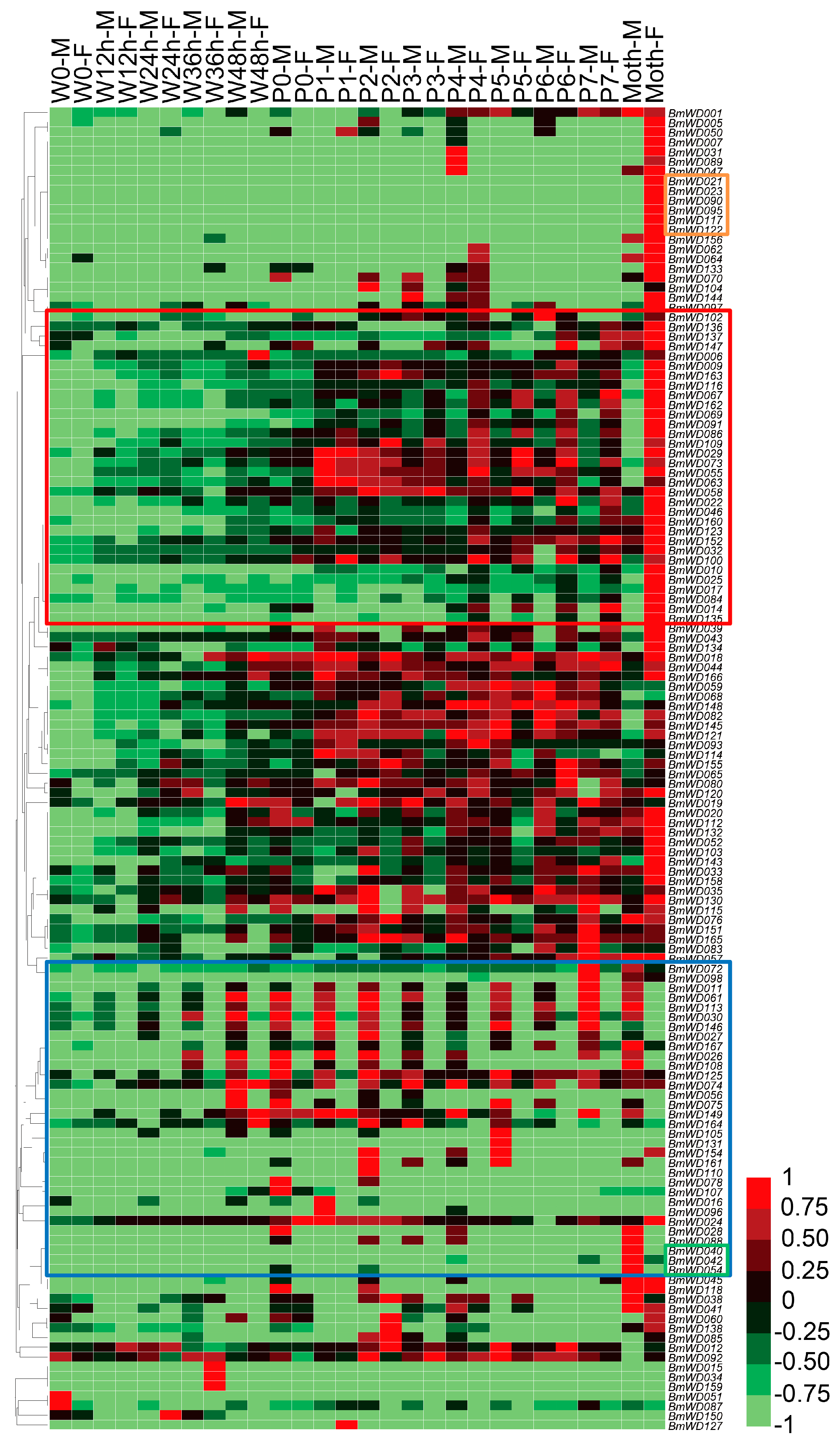
2.5. Spatial and Temporal Expression Profile of the BmWD40s
2.6. Hotspot Residues of Protein-Protein Interactions
3. Materials and Methods
3.1. Genome-Wide Identification of WD40 Proteins
3.2. Chromosomal Distribution and Gene Structure
3.3. Phylogenetic Analysis and Domain Architecture Annotation
3.4. Gene Ontology Annotation and Pathway Enrichment
3.5. Expression Profiling Using Microarray Data
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| WD | Tryptophan-Aspartic Acid (W-D) |
| LisH | Lis Homology |
| BEACH | Beige and Chediak–Higashi |
| CAF-1 | Chromatin Assembly Factor-1 |
| VLCFA | Very-Long-Chain Fatty Acids |
References
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.J.; Li, T.; Wang, Y.; Xiong, Y.; Wu, X.H.; Zhang, D.L.; Ye, Z.Q.; Wu, Y.D. Prokaryotic and Highly-Repetitive WD40 Proteins: A Systematic Study. Sci. Rep. 2017, 7, 10585. [Google Scholar] [CrossRef] [PubMed]
- Fong, H.K.; Hurley, J.B.; Hopkins, R.S.; Miake-Lye, R.; Johnson, M.S.; Doolittle, R.F.; Simon, M.I. Repetitive segmental structure of the transducin beta subunit: Homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. USA 1986, 83, 2162–2166. [Google Scholar] [CrossRef] [PubMed]
- Van der Voorn, L.; Ploegh, H.L. The WD-40 repeat. FEBS Lett. 1992, 307, 131–134. [Google Scholar] [CrossRef]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.A.; Coleman, D.E.; Lee, E.; Iniguez-Lluhi, J.A.; Posner, B.A.; Gilman, A.G.; Sprang, S.R. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 1995, 83, 1047–1058. [Google Scholar] [CrossRef]
- Lambright, D.G.; Sondek, J.; Bohm, A.; Skiba, N.P.; Hamm, H.E.; Sigler, P.B. The 2.0 A crystal structure of a heterotrimeric G protein. Nature 1996, 379, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Sondek, J.; Bohm, A.; Lambright, D.G.; Hamm, H.E.; Sigler, P.B. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature 1996, 379, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F. Diversity of WD-repeat proteins. Subcell. Biochem. 2008, 48, 20–30. [Google Scholar] [PubMed]
- Li, D.; Roberts, R. WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001, 58, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, M.; Kawashima, A.; Morioka, H.; Linn, S.; Sancar, A.; Mori, T.; Nikaido, O.; Matsunaga, T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2002, 277, 1637–1640. [Google Scholar] [CrossRef] [PubMed]
- Znaidi, S.; Pelletier, B.; Mukai, Y.; Labbe, S. The Schizosaccharomyces pombe corepressor Tup11 interacts with the iron-responsive transcription factor Fep1. J. Biol. Chem. 2004, 279, 9462–9474. [Google Scholar] [CrossRef] [PubMed]
- Jennings, B.H.; Pickles, L.M.; Wainwright, S.M.; Roe, S.M.; Pearl, L.H.; Ish-Horowicz, D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell 2006, 22, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Ajuh, P.; Sleeman, J.; Chusainow, J.; Lamond, A.I. A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem. 2001, 276, 42370–42381. [Google Scholar] [CrossRef] [PubMed]
- Vander Kooi, C.W.; Ren, L.; Xu, P.; Ohi, M.D.; Gould, K.L.; Chazin, W.J. The Prp19 WD40 domain contains a conserved protein interaction region essential for its function. Structure 2010, 18, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; McLean, J.R.; Hazbun, T.R.; Fields, S.; Vander Kooi, C.; Ohi, M.D.; Gould, K.L. Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS ONE 2011, 6, e16719. [Google Scholar] [CrossRef]
- Yan, C.; Hang, J.; Wan, R.; Huang, M.; Wong, C.C.; Shi, Y. Structure of a yeast spliceosome at 3.6—Angstrom resolution. Science 2015, 349, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Higa, L.A.; Wu, M.; Ye, T.; Kobayashi, R.; Sun, H.; Zhang, H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 2006, 8, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Garlick, J.D.; Kingston, R.E. Structural basis of histone H4 recognition by p55. Genes Dev. 2008, 22, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Suganuma, T.; Pattenden, S.G.; Workman, J.L. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 2008, 22, 1265–1268. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Cory, S. Apoptosomes: Engines for caspase activation. Curr. Opin. Cell Biol. 2002, 14, 715–720. [Google Scholar] [CrossRef]
- Yuan, S.; Yu, X.; Topf, M.; Ludtke, S.J.; Wang, X.; Akey, C.W. Structure of an apoptosome-procaspase-9 CARD complex. Structure 2010, 18, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Reubold, T.F.; Wohlgemuth, S.; Eschenburg, S. Crystal structure of full-length Apaf-1: How the death signal is relayed in the mitochondrial pathway of apoptosis. Structure 2011, 19, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.H.; Miettinen, P.J.; Maruoka, E.M.; Choy, L.; Derynck, R. A WD-domain protein that is associated with and phosphorylated by the type II TGF-beta receptor. Nature 1995, 377, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, R.; Bohm, A.; Sigler, P.B. Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin. Cell 1996, 87, 577–588. [Google Scholar] [CrossRef]
- Datta, P.K.; Moses, H.L. STRAP and Smad7 synergize in the inhibition of transforming growth factor beta signaling. Mol. Cell. Biol. 2000, 20, 3157–3167. [Google Scholar] [CrossRef] [PubMed]
- Gallenberger, M.; Meinel, D.M.; Kroeber, M.; Wegner, M.; Milkereit, P.; Bosl, M.R.; Tamm, E.R. Lack of WDR36 leads to preimplantation embryonic lethality in mice and delays the formation of small subunit ribosomal RNA in human cells in vitro. Hum. Mol. Genet. 2011, 20, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Koide, R.; Nakata, Y.; Saitsu, H.; Matsumoto, N.; Takahashi, K.; Nakano, I.; Orimo, S. A novel WDR45 mutation in a patient with static encephalopathy of childhood with neurodegeneration in adulthood (SENDA). Am. J. Med. Genet. A 2014, 164A, 2388–2390. [Google Scholar] [CrossRef] [PubMed]
- Bilguvar, K.; Ozturk, A.K.; Louvi, A.; Kwan, K.Y.; Choi, M.; Tatli, B.; Yalnizoglu, D.; Tuysuz, B.; Caglayan, A.O.; Gokben, S.; et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 2010, 467, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, A.K.; Khurshid, M.; Desir, J.; Carvalho, O.P.; Cox, J.J.; Thornton, G.; Kausar, R.; Ansar, M.; Ahmad, W.; Verloes, A.; et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010, 42, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Zhou, L.; Sun, W.; Zheng, Z.G.; Lu, P.; Gao, Y.; Yang, X.S.; Zhang, Z.C.; Tao, K.S.; et al. Fbxw7 regulates hepatocellular carcinoma migration and invasion via Notch1 signaling pathway. Int. J. Oncol. 2015, 47, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Wang, Y.; Zhao, S.; Liu, C.; Wang, Y.; Wen, M.; Mao, J.H.; Wei, G.; Zhang, P. FBXW7 negatively regulates ENO1 expression and function in colorectal cancer. Lab. Investig. 2015, 95, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Saez, B.; Mukhopadhyay, S.; Ding, D.; Ahmed, A.M.; Chen, X.; Pucci, F.; Yamin, R.; Wang, J.; Pittet, M.J.; et al. Tle1 tumor suppressor negatively regulates inflammation in vivo and modulates NF-kappaB inflammatory pathway. Proc. Natl. Acad. Sci. USA 2016, 113, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
- Orville Singh, C.; Xin, H.H.; Chen, R.T.; Wang, M.X.; Liang, S.; Lu, Y.; Cai, Z.Z.; Miao, Y.G. BmPLA2 containing conserved domain WD40 affects the metabolic functions of fat body tissue in silkworm, Bombyx mori. Insect Sci. 2016, 23, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Van Nocker, S.; Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Huang, X.; Lu, Z.; Yao, J. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genom. 2012, 13, 100. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhao, P.; Li, J.; Zhang, C.; Wang, L.; Ren, Z. Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Genet. Genom. 2014, 289, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Muthamilarasan, M.; Khan, Y.; Parida, S.K.; Prasad, M. Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e86852. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.D.; Hu, X.J.; Ma, J.; Li, T.; Ye, Z.Q.; Wu, Y.D. Genome-wide Analysis of WD40 Protein Family in Human. Sci. Rep. 2016, 6, 39262. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Schultz, J.; Milpetz, F.; Bork, P. SMART: Identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999, 27, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, X.J.; Zou, X.D.; Wu, X.H.; Ye, Z.Q.; Wu, Y.D. WDSPdb: A database for WD40-repeat proteins. Nucleic Acids Res. 2015, 43, D339–D344. [Google Scholar] [CrossRef] [PubMed]
- Angers, S.; Li, T.; Yi, X.; MacCoss, M.J.; Moon, R.T.; Zheng, N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006, 443, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant. Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, H.H.; Chung, W.Y.; Yang, J.; Li, W.H. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 2005, 22, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Shoja, V.; Zhang, L. A roadmap of tandemly arrayed genes in the genomes of human, mouse, and rat. Mol. Biol. Evol. 2006, 23, 2134–2141. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Takai, H.; Ogiwara, A.; Yokota, E.; Shimizu, T.; Inaba, K.; Mohri, H. Is outer arm dynein intermediate chain 1 multifunctional? Mol. Biol. Cell. 1996, 7, 1895–1907. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Harricane, M.C.; Lafitte, J.J.; Godard, P.; Zaegel, M.; Tack, V.; Lalau, G.; Bouvagnet, P. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome). Am. J. Hum. Genet. 2001, 68, 1030–1035. [Google Scholar] [CrossRef] [PubMed]
- Young, S.A.; Miyata, H.; Satouh, Y.; Kato, H.; Nozawa, K.; Isotani, A.; Aitken, R.J.; Baker, M.A.; Ikawa, M. CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis. Int. J. Mol. Sci. 2015, 16, 24732–24750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, H.; Li, W.; Zhang, Z.; Shang, X.; Zhang, D.; Li, Y.; Zhang, S.; Liu, J.; Hess, R.A.; et al. Intraflagellar transporter protein (IFT27), an IFT25 binding partner, is essential for male fertility and spermiogenesis in mice. Dev. Biol. 2017, 432, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Brites, P.; Mooyer, P.A.; El Mrabet, L.; Waterham, H.R.; Wanders, R.J. Plasmalogens participate in very-long-chain fatty acid-induced pathology. Brain 2009, 132, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.; Bayam, E.; Wagner, C.; Rinaldi, B.; Kretz, P.F.; Tilly, P.; Roos, M.; McGillewie, L.; Bar, S.; Minocha, S.; et al. WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proc. Natl. Acad. Sci. USA 2017, 114, E9308–E9317. [Google Scholar] [CrossRef] [PubMed]
- Dahlberg, C.; Kowalski, J.; Juo, P. WDR-20 and WDR-48 regulate the deubiquitinating enzyme USP-46 to promote the abundance of glutamate receptors in the ventral nerve cord of C. elegans. Mol. Biol. Cell. 2013, 299, 3444–3456. [Google Scholar]
- Lodder, E.M.; De Nittis, P.; Koopman, C.D.; Wiszniewski, W.; Moura de Souza, C.F.; Lahrouchi, N.; Guex, N.; Napolioni, V.; Tessadori, F.; Beekman, L.; et al. GNB5 Mutations Cause an Autosomal-Recessive Multisystem Syndrome with Sinus Bradycardia and Cognitive Disability. Am. J. Hum. Genet. 2016, 99, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Huang, J.D.; Mermall, V.; Hamilton, B.A.; Mooseker, M.S.; Escayg, A.; Copeland, N.G.; Jenkins, N.A.; Meisler, M.H. The mouse neurological mutant flailer expresses a novel hybrid gene derived by exon shuffling between Gnb5 and Myo5a. Hum. Mol. Genet. 2000, 9, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Kottgen, A.; Pattaro, C.; Boger, C.A.; Fuchsberger, C.; Olden, M.; Glazer, N.L.; Parsa, A.; Gao, X.; Yang, Q.; Smith, A.V.; et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010, 42, 376–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [PubMed]
- Zhao, Q.; Zhu, Z.; Kasahara, M.; Morishita, S.; Zhang, Z. Segmental duplications in the silkworm genome. BMC Genom. 2013, 14, 521. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Cheng, D.; Duan, J.; Wang, G.; Cheng, T.; Zha, X.; Liu, C.; Zhao, P.; Dai, F.; Zhang, Z.; et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007, 8, R162. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Qian, W.; Meng, M.; Wang, Y.; Peng, J.; Xia, Q. Identification and Expression Profiling of the BTB Domain-Containing Protein Gene Family in the Silkworm, Bombyx mori. Int. J. Genom. 2014, 2014, 865065. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Tong, X.; Han, M.; Hu, H.; Dai, F. Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 527. https://doi.org/10.3390/ijms19020527
He S, Tong X, Han M, Hu H, Dai F. Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx mori. International Journal of Molecular Sciences. 2018; 19(2):527. https://doi.org/10.3390/ijms19020527
Chicago/Turabian StyleHe, Songzhen, Xiaoling Tong, Minjin Han, Hai Hu, and Fangyin Dai. 2018. "Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx mori" International Journal of Molecular Sciences 19, no. 2: 527. https://doi.org/10.3390/ijms19020527




