Functional Insights into the Roles of Hormones in the Dendrobium officinale-Tulasnella sp. Germinated Seed Symbiotic Association
Abstract
:1. Introduction
2. Results
2.1. Detecting Mycorrhizal Fungus Colonization in Germinated Seeds of D. officinale
2.2. Transcriptome Sequence Assembly and Annotation.
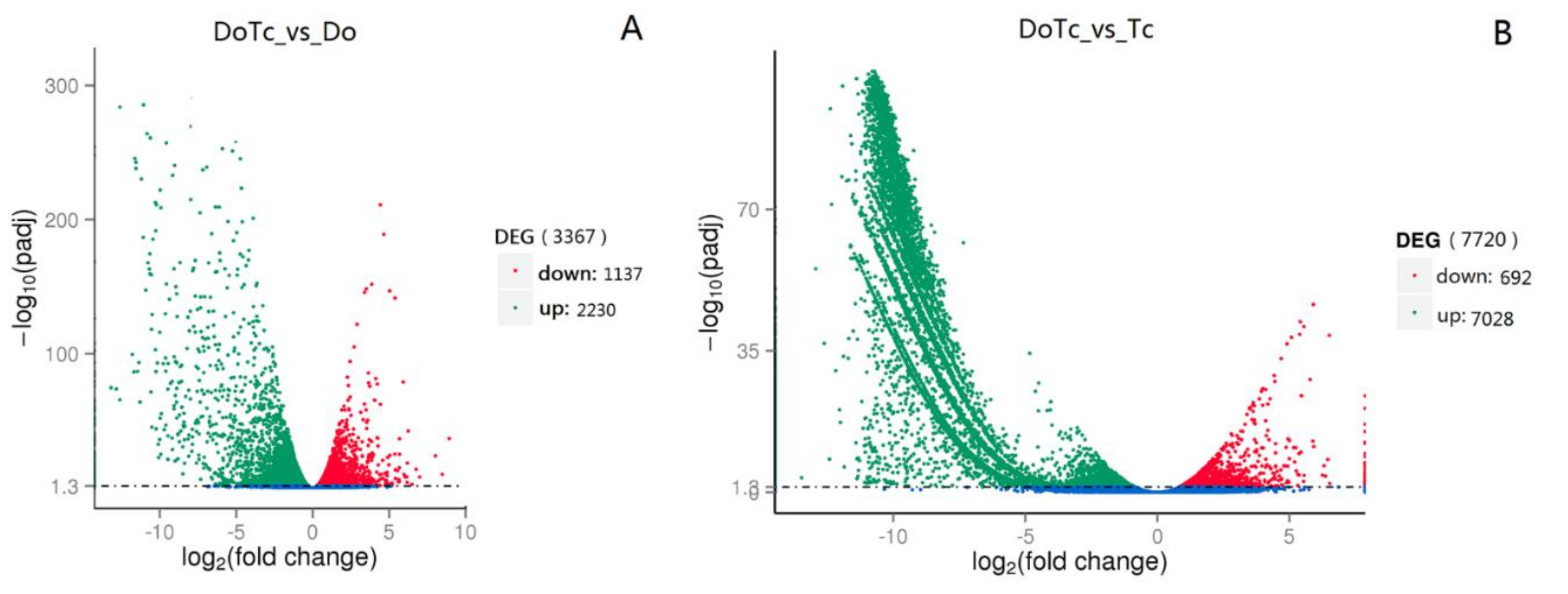
2.3. Differentially Expressed Genes in Symbiotic and Asymbiotic Germinated Seeds and Mycorrhizal Fungi
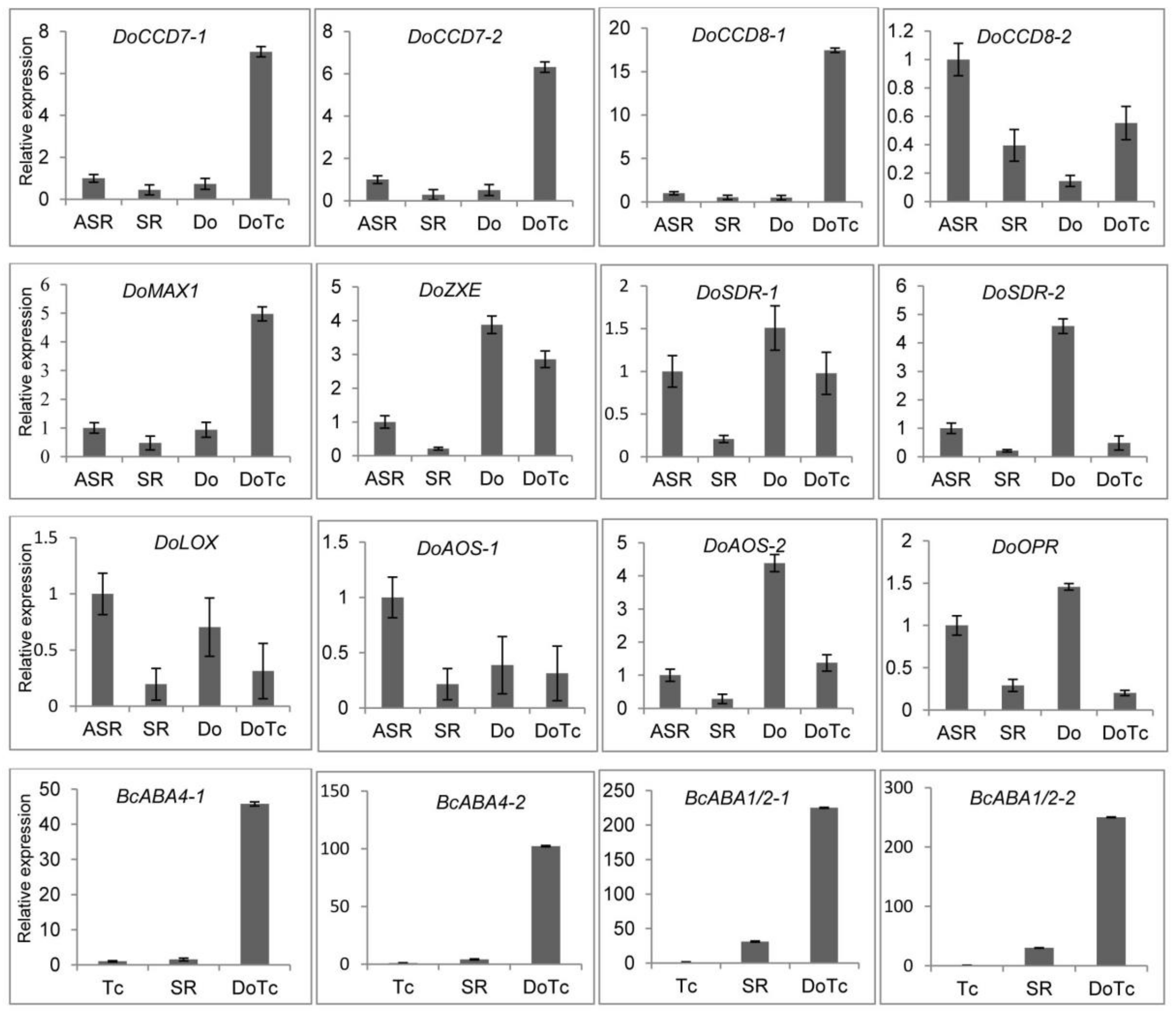
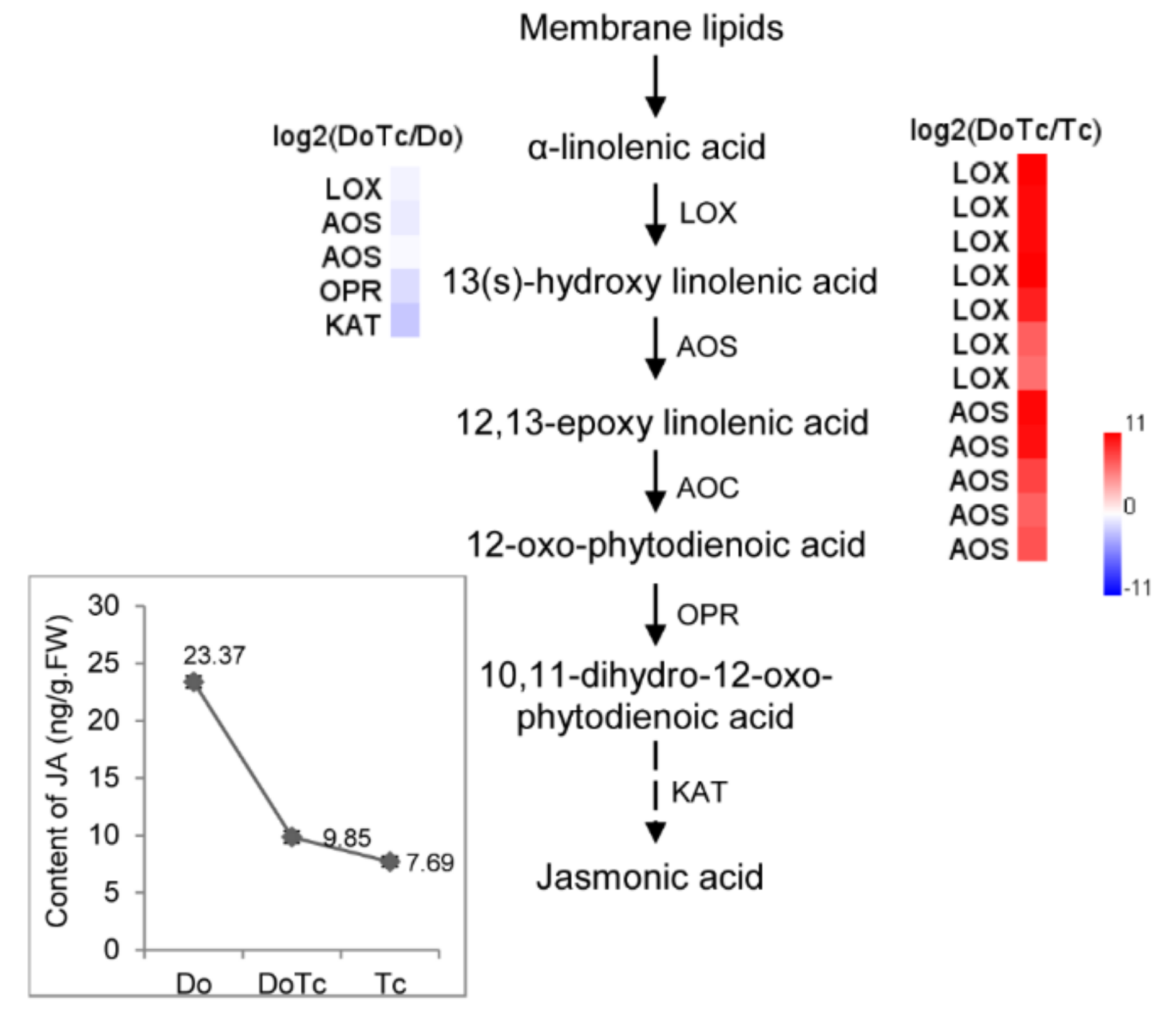
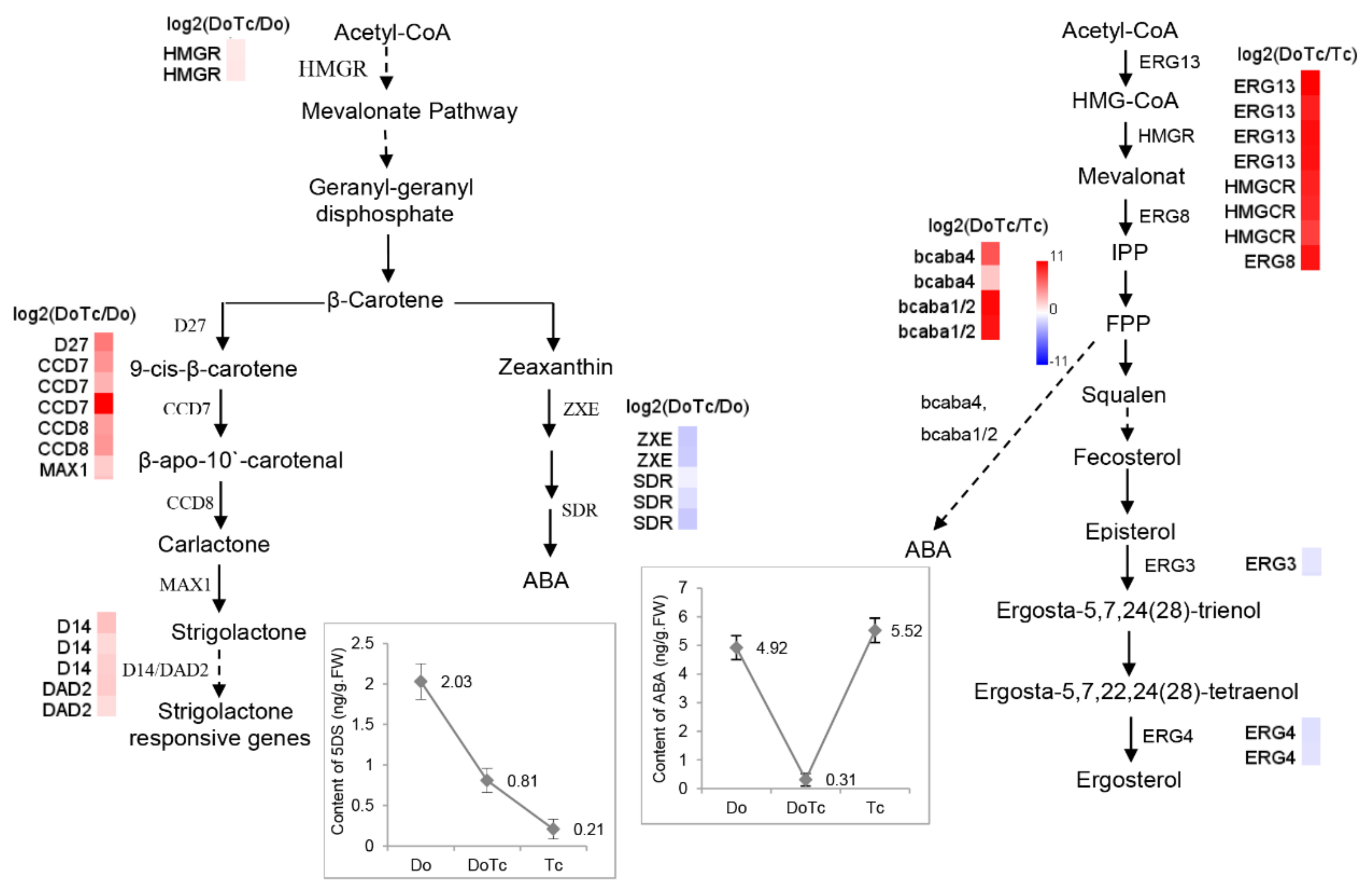
2.4. Expression of DEGs Related to ABA, JA, and SL Hormone Biosynthesis Detected by qRT-PCR
2.5. Assays of ABA, JA, and SL Contents
3. Discussion
4. Materials and Methods
4.1. Symbiotic and Asymbiotic Germination of D. officinale Seeds
4.2. cDNA Library Construction and Nucleotide Sequencing
4.3. Transcriptome Data Analysis
4.3.1. Quality Control and cDNA Assembly
4.3.2. Quantification of Gene Expression Levels
4.3.3. Differential Gene Expression Analysis
4.3.4. Unigene Functional Annotation and Metabolic Pathway Analysis
4.4. Hormone Extraction, Fractionation, and UPLC-ESI-qMS/MS Analysis
4.5. Differential Gene Expression Detected by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arditti, J. Factors affecting the germination of orchid seeds. Bot. Rev. 1967, 33, 1–97. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, T.J.A.; Tsavkelova, E.A.; Zeng, S.; Ng, T.B.; Parthibhan, S.; Dobranszki, J.; Cardoso, J.C.; Rao, M.V. Symbiotic in vitro seed propagation of Dendrobium: Fungal and bacterial partners and their influence on plant growth and development. Planta 2015, 242, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lu, C.; Chung, M.; Yeung, E.C.; Lee, N. Developmental changes in endogenous abscisic acid concentrations and asymbiotic seed germination of a terrestrial orchid, Calanthe tricarinata Lindl. J. Am. Soc. Hortc. Sci. 2007, 132, 246–252. [Google Scholar]
- Liu, H.; Luo, Y.; Liu, H. Studies of mycorrhizal fungi of Chinese orchids and their role in orchid conservation in China: A review. Bot. Rev. 2010, 76, 241–262. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, G.; Zhang, D.; Hsiao, Y.; Guo, S. ESTs analysis reveals putative genes involved in symbiotic seed germination in Dendrobium officinale. PLoS ONE 2013, 8, e72705. [Google Scholar] [CrossRef] [PubMed]
- Valadares, R.B.S.; Perotto, S.; Santos, E.C.; Lambais, M.R. Proteome changes in Oncidium sphacelatum (Orchidaceae) at different trophic stages of symbiotic germination. Mycorrhiza 2014, 24, 349–360. [Google Scholar] [CrossRef] [PubMed]
- López-Chávez, M.Y.; Guillén-Navarro, K.; Bertolini, V.; Encarnación, S.; Hernández-Ortiz, M.; Sánchez-Moreno, I.; Damon, A. Proteomic and morphometric study of the in vitro interaction between Oncidium sphacelatum Lindl. (Orchidaceae) and Thanatephorus sp. RG26 (Ceratobasidiaceae). Mycorrhiza 2016, 26, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Miura, C.; Yamaguchi, K.; Miyahara, R.; Yamamoto, T.; Fuji, M.; Yagame, T.; Imaizumi-Anraku, H.; Yamato, M.; Shigenobu, S.; Kaminaka, H. The mycoheterotrophic symbiosis between orchids and mycorrhizal fungi possesses major components shared with mutualistic plant-mycorrhizal symbioses. Mol. Plant Microbe Interact. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Wu, J.R.; Duan, X.T.; Qin, H.X. Plant hormones produced from symbionts of Cymbidium goeringii and four Rhizoctonia-like strains. Chin. Guizhou Agric. Sci. 2017, 45, 84–86. [Google Scholar] [CrossRef]
- Chen, J.; Liu, S.S.; Kohler, A.; Yan, B.; Luo, H.M.; Chen, X.M.; Guo, S.X. iTRAQ and RNA-seq analyses provide new insights into regulation mechanism of symbiotic germination of Dendrobium officinale seeds (Orchidaceae). J. Proteome Res. 2017, 16, 2174–2187. [Google Scholar] [CrossRef] [PubMed]
- Chanclud, E.; Morel, J. Plant hormones: A fungal point of view. Mol. Plant Pathol. 2016, 17, 1289–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, S.L.; Zettler, L.W. Symbiotic germination of three semiaquatic rein orchids (Habenaria repens, H. quinquiseta, H. macroceratitis) from florida. Aquatic Bot. 2002, 72, 25–35. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Ann. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Oritani, T.; Kiyota, H. Biosynthesis and metabolism of abscisic acid and related compounds. Nat. Prod. Rep. 2003, 20, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Shu, D.; Yang, J.; Ding, Z.T.; Tan, H. Sequencing and transcriptional analysis of the biosynthesis gene cluster of abscisic acid-producing Botrytis cinerea. Int. J. Mol. Sci. 2014, 15, 17396–17410. [Google Scholar] [CrossRef] [PubMed]
- Lievens, L.; Pollier, J.; Goossens, A.; Beyaert, R.; Staal, J. Abscisic acid as pathogen effector and immune regulator. Front. Plant Sci. 2017, 8, 587. [Google Scholar] [CrossRef] [PubMed]
- Siewers, V.; Smedsgaard, J.; Tudzynski, P. The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl. Environ. Microb. 2004, 70, 3868–3876. [Google Scholar] [CrossRef] [PubMed]
- Stec, N.; Banasiak, J.; Jasiński, M. Abscisic acid: An overlooked player in plant–microbe symbioses formation. Acta Biochim. Pol. 2016, 63, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Hirai, N.; Yoshida, R.; Todoroki, Y.; Ohigashi, H. Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotech. Biochem. 2000, 64, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhang, Z.; Zhong, J.; Di, L.; Zhou, J.; Yang, J.; Xiao, L.; Shu, D.; Tan, H. Comparative transcriptome analysis between an evolved abscisic acid-overproducing mutant Botrytis cinerea TBC-A and its ancestral strain Botrytis cinerea TBC-6. Sci. Rep. 2016, 6, 37487–37499. [Google Scholar] [CrossRef] [PubMed]
- Zdyb, A.; Salgado, M.G.; Demchenko, K.N.; Brenner, W.G.; Płaszczyca, M.; Stumpe, M.; Herrfurth, C.; Feussner, I.; Pawlowski, K. Allene oxide synthase, allene oxide cyclase and jasmonic acid levels in Lotus japonicus nodules. PLoS ONE 2018, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Liu, J.; Liu, J.; Yang, X.; Chen, X.; Gu, M.; Gu, A. Advances in the response and modulation of phytohormones on arbuscular mycorrhizal symbiosis. J. Plant Nutr. Fert. 2016, 22, 1679–1689. [Google Scholar] [CrossRef]
- Prandi, C.; Cardinale, F. Strigolactones: A New Class of Plant Hormones with Multifaceted Roles; John Wiley & Sons: Chichester, UK, 2014. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Cao, M.; Akiyama, K.; Akatsu, T.; Seto, Y.; Hanada, A.; Li, W.; Takeda-Kamiya, N.; Morimoto, Y.; Yamaguchi, S. Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 2015, 56, 1059–1072. [Google Scholar] [CrossRef] [PubMed]
- Saeed, W.; Naseem, S.; Ali, Z. Strigolactones biosynthesis and their role in abiotic stress resilience in plants: A critical review. Front. Plant Sci. 2017, 8, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, G.; Gui, R.; Zhang, H.; Hu, X. Determination of strigolactones extracted from root of Phyllostachys edulis by ultra performance liquid chromatography. J. Zhejiang Forest. Coll. 2013, 30, 607–610. [Google Scholar]
- Walker-Simmons, M. ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 1987, 84, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Pence, V.C. Abscisic acid and the maturation of cacao embryos in vitro. Plant Physiol. 1992, 98, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Crouch, M.L. Rapeseed embryo development in culture on high osmoticum is similar to that in seeds. Plant Physiol. 1986, 81, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Van, K.G. Abscisic acid in terrestrial orchid seeds: A possible impact on their germination. Lindleyana 1987, 2, 84–87. [Google Scholar]
- Wu, H.; Song, X.; Yang, S.; Wang, J. Effects of symbiotic fungi on the physiological characteristics of Dendrobium catenatum Lindley (Orchidaceae). Chin. Plant Sci. J. 2011, 29, 738–742. [Google Scholar] [CrossRef]
- Chen, J.; Wang, H.; Liu, S.S.; Li, Y.Y.; Guo, S.X. Ultrastructure of symbiotic germination of the orchid Dendrobium officinale with its mycobiont, Sebacina sp. Aust. J. Bot. 2014, 62, 229–234. [Google Scholar] [CrossRef]
- Umezawa, T.; Okamoto, M.; Kushiro, T.; Nambara, E.; Oono, Y.; Kobayashi, M.S.M.; Koshiba, T.; Kamiya, Y.; Shinozaki, K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006, 46, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Zeevaart, J.A.D.; Creelman, R.A. Metabolism and physiology of abscisic acid. Annu. Rev. Plant Biol. 1988, 39, 439–473. [Google Scholar] [CrossRef]
- Lopez-Raez, J.A.; Kohlen, W.; Charnikhova, T.; Mulder, P.; Undas, A.K.; Sergeant, M.J.; Verstappen, F.; Bugg, T.D.; Thompson, A.J.; Ruyter-Spira, C.; et al. Does abscisic acid affect strigolactone biosynthesis? New Phytol. 2010, 187, 343–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toh, S.; Kamiya, Y.; Kawakami, N.; Nambara, E.; McCourt, P.; Tsuchiya, Y. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol. 2012, 53, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Tian, J.; Wang, T.; Li, L. Symbiosis established between orchid and Tulasnella spp. fungi. J. Nucl. Agric. Sci. 2017, 31, 876–883. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, J.; Liu, S.; Chen, C.; Zhu, H.; Cao, J. The colonization patterns of different fungi on roots of Cymbidium hybridum plantlets and their respective inoculation effects on growth and nutrient uptake of orchid plantlets. World J. Microbiol. Biotech. 2014, 30, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Xian, A.L.F.; Raychowdhury, R.; Zeng, Q.; Chen, Z.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2011, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, J.; Li, S.; Zeng, X.; Meng, Z.; Guo, S. Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Orchidaceae). Int. J. Mol. Sci. 2015, 16, 30190–30203. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.; Sun, L.; Xu, X.; Zhu, B.; Xu, H. Differential expression of VvLOXA diversifies C6 volatile profiles in some Vitis vinifera table grape cultivars. Int. J. Mol. Sci. 2017, 18, 2705–2719. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Zhu, Q.; Pei, W.; Fan, J.; Liang, Y.; Cui, Y.; Lv, N.; Wang, W. Whole-transcriptome selection and evaluation of internal reference genes for expression analysis in protocorm development of Dendrobium officinale Kimura et Migo. PLoS ONE 2016, 11, e0163478. [Google Scholar] [CrossRef] [PubMed]
- Fochi, V.; Chitarra, W.; Kohler, A.; Voyron, S.; Singan, V.R.; Lindquist, E.A.; Barry, K.W.; Girlanda, M.; Grigoriev, I.V.; Martin, F.; et al. Fungal and plant gene expression in the Tulasnella calospora–Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytol. 2017, 213, 365–379. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Description | |
|---|---|---|---|
| ABA pathway | log2(DoTc/Do) | ||
| DoZXE-1 | Dendrobium_GLEAN_10076665 | −2.11 | Zeaxanthin epoxidase |
| DoZXE-2 | Novel00252 | −2.00 | Zeaxanthin epoxidase |
| DoSDR-1 | Dendrobium_GLEAN_10042288 | −0.57 | Short-chain type dehydrogenase/reductase |
| DoSDR-2 | Dendrobium_GLEAN_10068100 | −1.33 | Short-chain type dehydrogenase/reductase |
| DoSDR-3 | Dendrobium_GLEAN_10127951 | −2.1 | Short-chain type dehydrogenase/reductase |
| ABA pathway | log2(DoTc/Tc) | ||
| BcABA4-1 | Cluster-2641.12008 | 7.24 | Short-chain type dehydrogenase/reductase dehydrogenase/reductase |
| BcABA4-2 | Cluster-2152.0 | 2.42 | Short-chain type dehydrogenase/reductase |
| BcABA1/2-1 | Cluster-2641.19250 | 10.43 | P450 monooxygenase protein CYP51 |
| BcABA1/2-2 | Cluster-2641.15199 | 10.01 | P450 monooxygenase protein CYP51 |
| JA pathway | log2(DoTc/Do) | ||
| DoLOX | Dendrobium_GLEAN_10090503 | −0.67 | Linoleate 13S-lipoxygenase |
| DoAOS-1 | Dendrobium_GLEAN_10010006 | −0.98 | Allene oxide synthase |
| DoAOS-2 | Dendrobium_GLEAN_10012197 | −0.40 | Allene oxide synthase |
| DoOPR | Dendrobium_GLEAN_10113554 | −1.65 | 12-oxophytodienoate reductase |
| DoKAT | Dendrobium_GLEAN_10097185 | −2.50 | 3-ketoacyl-CoA thiolase |
| JA pathway | log2(DoTc/Tc) | ||
| TcLOX-1 | Cluster-2641.17948 | 10.60 | Linoleate 13S-lipoxygenase |
| TcLOX-2 | Cluster-2641.19486 | 10.33 | Linoleate 13S-lipoxygenase |
| TcLOX-3 | Cluster-2641.18695 | 10.34 | Linoleate 13S-lipoxygenase |
| TcLOX-4 | Cluster-2641.17988 | 10.66 | Linoleate 13S-lipoxygenase |
| TcLOX-5 | Cluster-2641.16009 | 9.35 | Linoleate 13S-lipoxygenase |
| TcLOX-6 | Cluster-6210.0 | 6.67 | Linoleate 13S-lipoxygenase |
| TcLOX-7 | Cluster-1378.1 | 5.99 | Linoleate 13S-lipoxygenase |
| TcAOS-1 | Cluster-2641.19289 | 10.40 | Allene oxide synthase |
| TcAOS-2 | Cluster-2641.20726 | 10.07 | Allene oxide synthase |
| TcAOS-3 | Cluster-2641.16940 | 7.81 | Allene oxide synthase |
| TcAOS-4 | Cluster-2641.8055 | 6.54 | Allene oxide synthase |
| TcAOS-5 | Cluster-2641.30606 | 7.21 | Allene oxide synthase |
| SL pathway | log2(DoTc/Do) | ||
| DoHMGR-1 | Dendrobium_GLEAN_10079864 | 0.87 | Hydroxymethylglutaryl-CoA reductase |
| DoHMGR-2 | Dendrobium_GLEAN_10079865 | 0.91 | Hydroxymethylglutaryl-CoA reductase |
| DoD27 | Dendrobium_GLEAN_10126990 | 5.22 | β-carotene isomerase |
| DoCCD7-1 | Dendrobium_GLEAN_10008301 | 4.21 | Carotenoid cleavage dioxygenase 7 |
| DoCCD7-2 | Dendrobium_GLEAN_10009315 | 2.98 | Carotenoid cleavage dioxygenase 7 |
| DoCCD7-3 | Dendrobium_GLEAN_10049685 | 9.92 | Carotenoid cleavage dioxygenase 7 |
| DoCCD8-1 | Dendrobium_GLEAN_10070249 | 3.87 | Carotenoid cleavage dioxygenase 8 |
| DoCCD8-2 | Dendrobium_GLEAN_10072088 | 4.08 | Carotenoid cleavage dioxygenase 8 |
| DoMAX1 | Dendrobium_GLEAN_10109899 | 2.00 | Cytochrome P450 711A1 |
| DoD14-1 | Dendrobium_GLEAN_10025168 | 2.45 | Probable strigolactone esterase |
| DoD14-2 | Dendrobium_GLEAN_10114182 | 1.52 | Probable strigolactone esterase |
| DoD14-3 | Dendrobium_GLEAN_10143398 | 1.86 | Probable strigolactone esterase |
| DoDAD2-1 | Dendrobium_GLEAN_10013491 | 2.01 | Probable strigolactone esterase |
| DoDAD2-2 | Dendrobium_GLEAN_10114181 | 1.34 | Probable strigolactone esterase |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Song, Z.; Wang, X.; Xu, L.; Sun, Q.; Li, L. Functional Insights into the Roles of Hormones in the Dendrobium officinale-Tulasnella sp. Germinated Seed Symbiotic Association. Int. J. Mol. Sci. 2018, 19, 3484. https://doi.org/10.3390/ijms19113484
Wang T, Song Z, Wang X, Xu L, Sun Q, Li L. Functional Insights into the Roles of Hormones in the Dendrobium officinale-Tulasnella sp. Germinated Seed Symbiotic Association. International Journal of Molecular Sciences. 2018; 19(11):3484. https://doi.org/10.3390/ijms19113484
Chicago/Turabian StyleWang, Tao, Zheng Song, Xiaojing Wang, Lijun Xu, Qiwu Sun, and Lubin Li. 2018. "Functional Insights into the Roles of Hormones in the Dendrobium officinale-Tulasnella sp. Germinated Seed Symbiotic Association" International Journal of Molecular Sciences 19, no. 11: 3484. https://doi.org/10.3390/ijms19113484





