The Mitochondrial Genes BAK1, FIS1 and SFN are Linked with Alterations in Mitochondrial Membrane Potential in Barrett’s Esophagus
Abstract
:1. Introduction
2. Results
2.1. In Vitro Screening Using Human PCR Mitochondrial Gene Microarrays
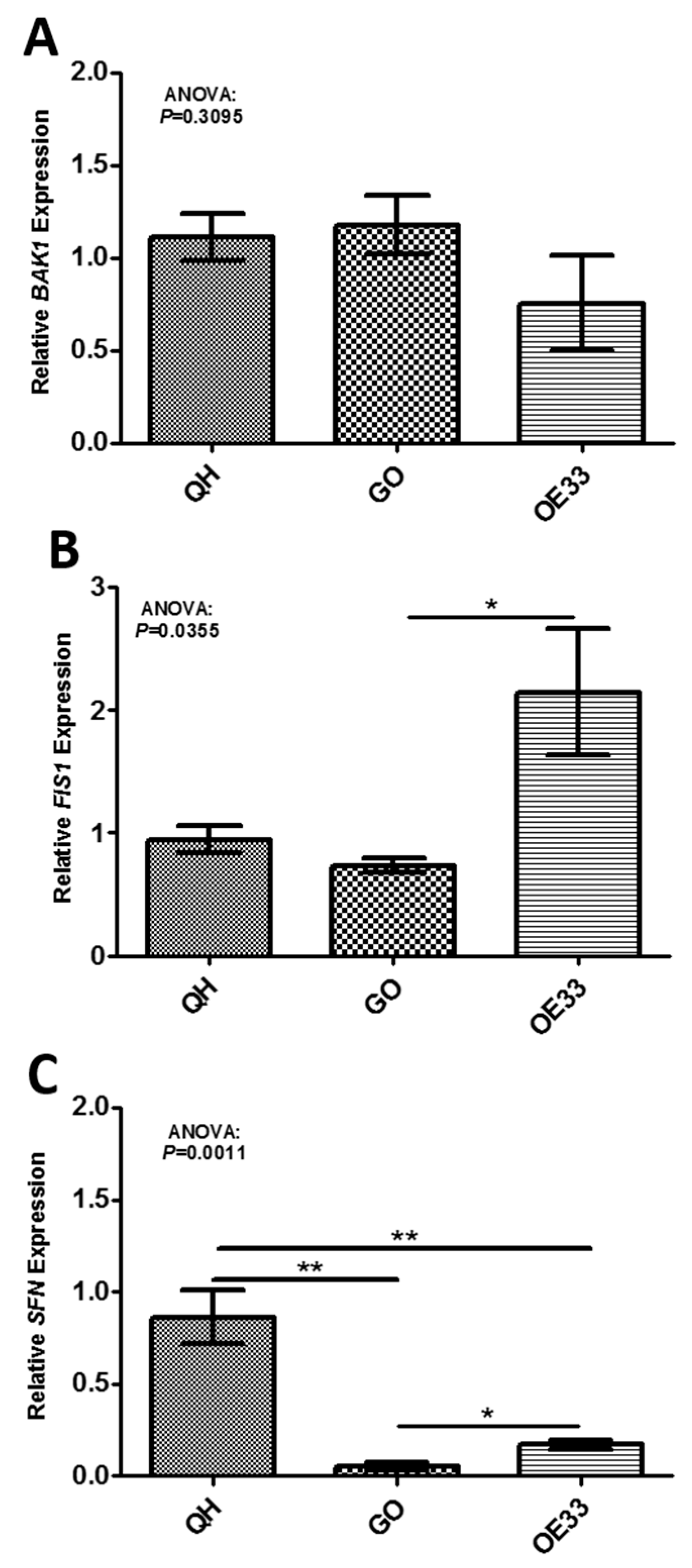
2.2. In Vivo Validation of Gene Targets
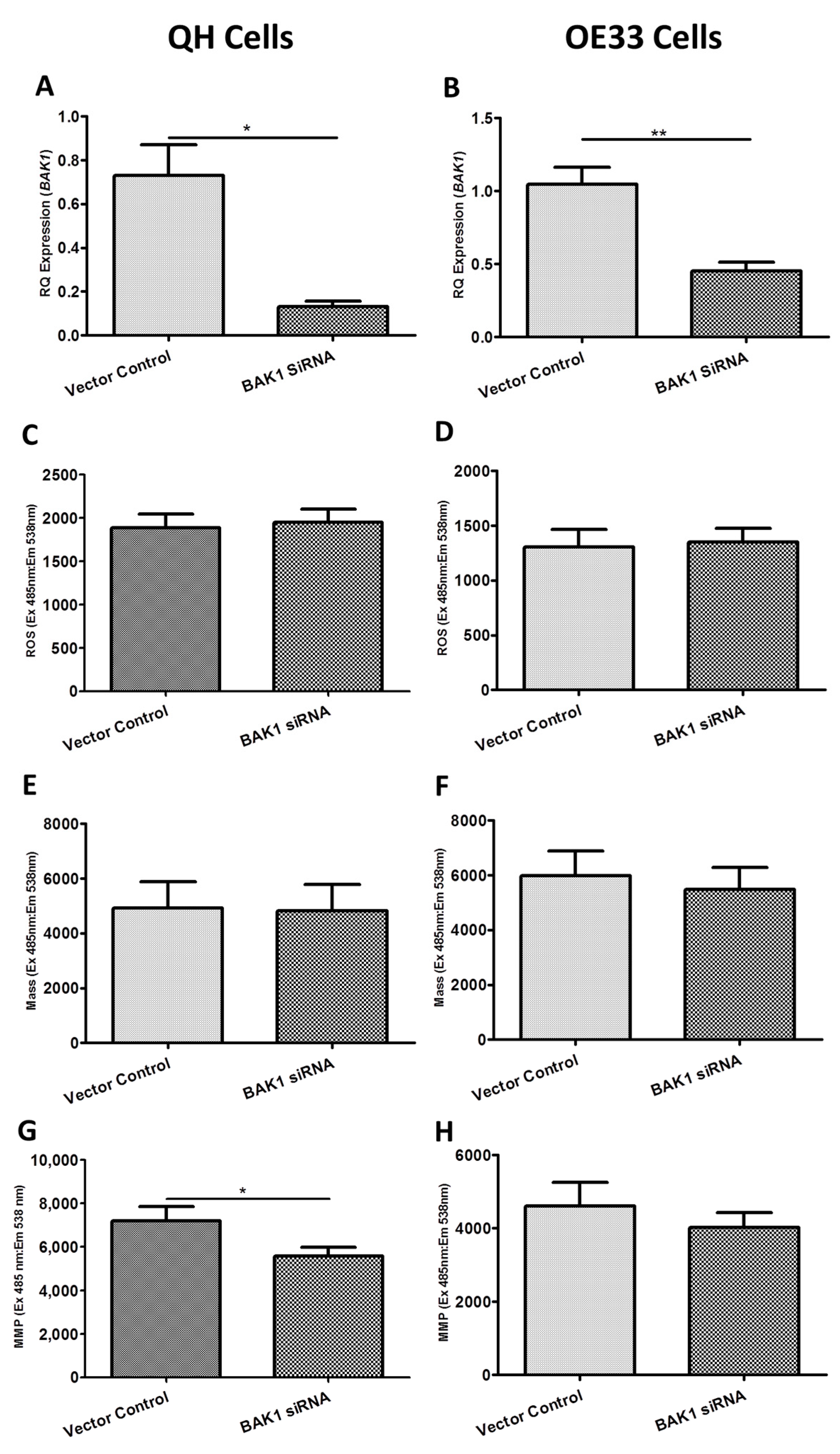
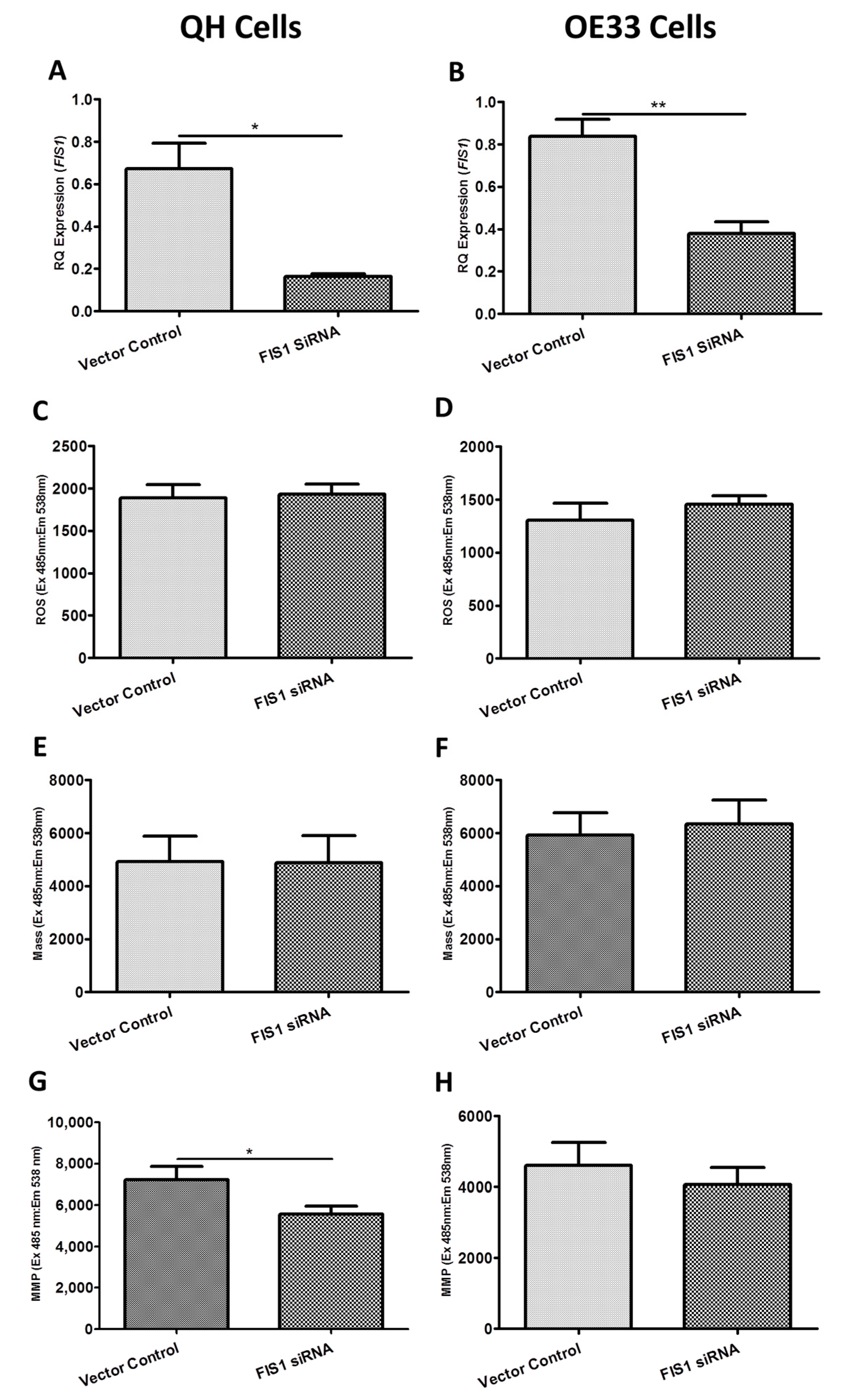
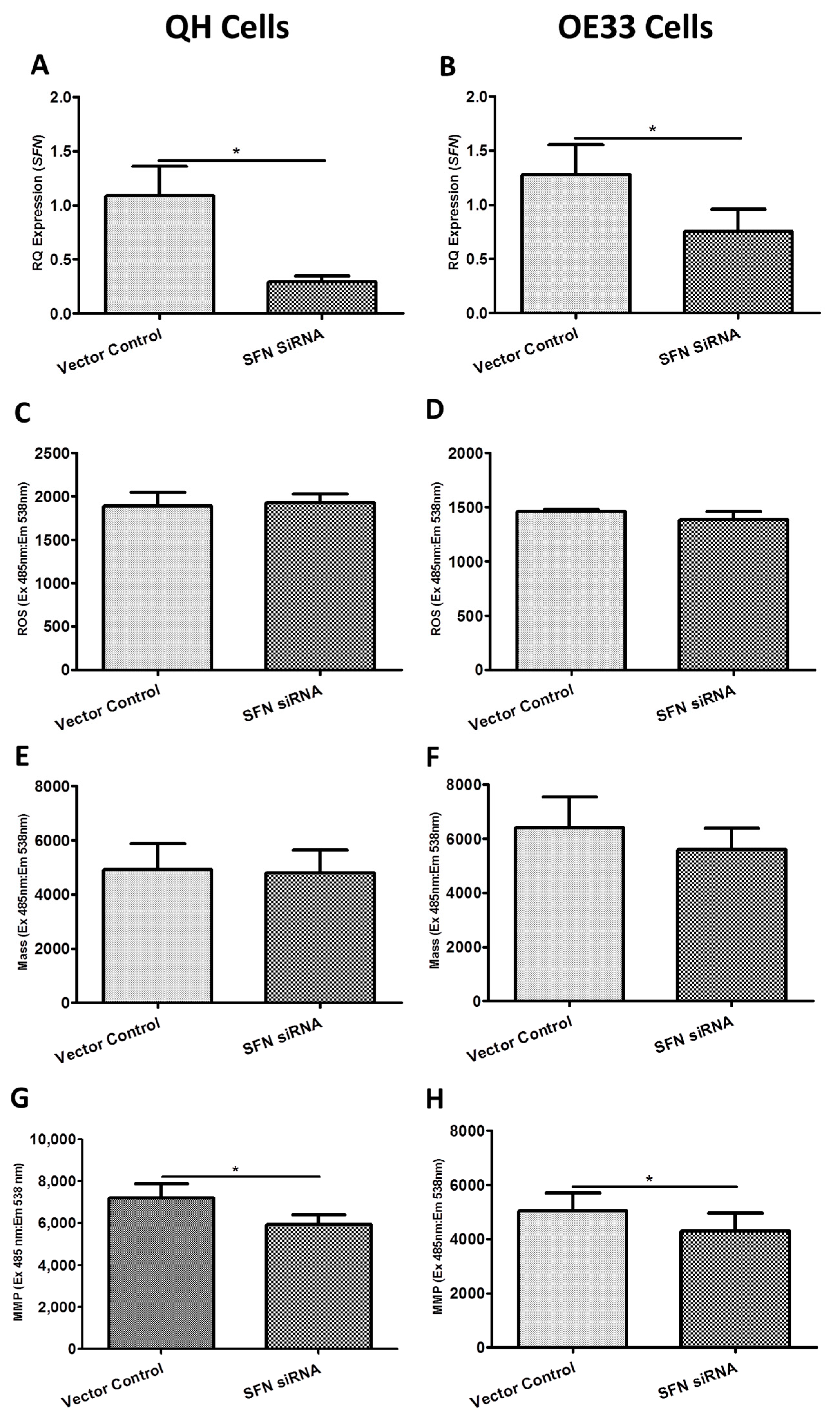
2.3. Functional Effect of BAK1, FIS1 and SFN siRNA Knockdown on Reactive Oxygen Species (ROS) Production, Mitochondrial Mass and Mitochondrial Membrane Potential (MMP) In Vitro
2.4. Assessing the Effect of BAK1, FIS1 and SFN siRNA Knockdown on Cellular Metabolism In Vitro
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Metaplastic, Dysplastic and Adenocarcinogenic Cell Lines
4.3. Screening via qRT-PCR Microarray Analysis
4.4. In Vitro qRT-PCR Validation of Gene Targets
4.5. In Vivo qRT-PCR Validation of Gene Targets
4.6. In Vitro siRNA Knockdown of Gene Targets
4.7. Exploring the Functional Effect of siRNA Knockdown on Reactive Oxygen Species Production, Mitochondrial Mass and Mitochondrial Membrane Potential In Vitro
4.8. Characterizing the Metabolic Effect of siRNA Knockdown of BAK1, FIS1 and SFN Utilizing the Seahorse XFE24 Analyzer In Vitro
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jankowski, J.; Barr, H.; Wang, K.; Delaney, B. Diagnosis and management of Barrett’s oesophagus. BMJ 2010, 341, c4551. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.V.; Donohoe, C.L.; McGillycuddy, E.; Ravi, N.; O’Toole, D.; O’Byrne, K.; Hollywood, D. Evolving progress in oncologic and operative outcomes for esophageal and junctional cancer: Lessons from the experience of a high-volume center. J. Thoracic. Cardiov. Surg. 2012, 143, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.N.; Czajka, A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 2013, 13, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Brahimi-Horn, M.C.; Mazure, N.M. Hypoxic VDAC1: A potential mitochondrial marker for cancer therapy. Adv. Exp. Med. Biol. 2014, 772, 101–110. [Google Scholar] [PubMed]
- Jakupciak, J.P.; Dakubo, G.D.; Maragh, S.; Parr, R.L. Analysis of potential cancer biomarkers in mitochondrial DNA. Curr. Opin. Mol. Ther. 2006, 8, 500–506. [Google Scholar] [PubMed]
- Amoedo, N.D.; Rodrigues, M.F.; Rumjanek, F.D. Mitochondria: Are mitochondria accessory to metastasis? Int. J. Biochem. Cell Biol. 2014, 51, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Whitaker-Menezes, D.; Martinez-Outschoorn, U.E.; Salem, A.F.; Tsirigos, A.; Lamb, R.; Sneddon, S.; Hulit, J.; Howell, A.; Lisanti, M.P. Mitochondria “fuel” breast cancer metabolism: Fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle 2012, 11, 4390–4401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandary, B.; Marahatta, A.; Kim, H.R.; Chae, H.J. Mitochondria in relation to cancer metastasis. J. Bioenerg. Biomembr. 2012, 44, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Phelan, J.J.; MacCarthy, F.; Feighery, R.; O’Farrell, N.J.; Lynam-Lennon, N.; Doyle, B.; O’Toole, D.; Ravi, N.; Reynolds, J.V.; O’Sullivan, J. Differential expression of mitochondrial energy metabolism profiles across the metaplasia-dysplasia-adenocarcinoma disease sequence in Barrett’s oesophagus. Cancer Lett. 2014, 354, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, S.; Venkatraman, A.; Ramakrishna, B.S. Impairment of mitochondrial acetoacetyl CoA thiolase activity in the colonic mucosa of patients with ulcerative colitis. Gut 2007, 56, 1543–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sifroni, K.G.; Damiani, C.R.; Stoffel, C.; Cardoso, M.R.; Ferreira, G.K.; Jeremias, I.C.; Rezin, G.T.; Scaini, G.; Schuck, P.F.; Dal-Pizzol, F.; et al. Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol. Cell. Biochem. 2010, 342, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.R.; Benetton, C.A.; Stoffel, C.; Bardini, K.C.; Cardoso, V.H.; di Giunta, G.; Pinho, R.A.; Dal-Pizzol, F.; Streck, E.L. Oxidative stress and metabolism in animal model of colitis induced by dextran sulfate sodium. J. Gastroenterol. Hepatol. 2007, 22, 1846–1851. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, S.; Rajamanickam, S.; Motamarry, A.; Ramakrishna, B.S.; Amirtharaj, J.G.; Ramachandran, A.; Pulimood, A.; Venkatraman, A. Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel Dis. 2012, 18, 2158–2168. [Google Scholar] [CrossRef] [PubMed]
- Ussakli, C.H.; Ebaee, A.; Binkley, J.; Brentnall, T.A.; Emond, M.J.; Rabinovitch, P.S.; Risques, R.A. Mitochondria and tumor progression in ulcerative colitis. J. Natl. Cancer Inst. 2013, 105, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Brentnall, T.A.; Pan, S.; Bronner, M.P.; Crispin, D.A.; Mirzaei, H.; Cooke, K.; Tamura, Y.; Nikolskaya, T.; Jebailey, L.; Goodlett, D.R.; et al. Proteins That Underlie Neoplastic Progression of Ulcerative Colitis. Proteom. Clin. Appl. 2009, 3, 1326. [Google Scholar] [CrossRef]
- Ostuni, M.A.; Issop, L.; Peranzi, G.; Walker, F.; Fasseu, M.; Elbim, C.; Papadopoulos, V.; Lacapere, J.J. Overexpression of translocator protein in inflammatory bowel disease: Potential diagnostic and treatment value. Inflamm. Bowel Dis. 2010, 16, 1476–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenner, C.E. Targeting mitochondria as a therapeutic target in cancer. J. Cell. Physiol. 2012, 227, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.S.; Zheng, L.D.; Wang, L.; Liu, J.; Qian, W. BAK overexpression mediates p53-independent apoptosis inducing effects on human gastric cancer cells. BMC Cancer 2004, 4, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lugering, A.; Schmidt, M.; Lugering, N.; Pauels, H.G.; Domschke, W.; Kucharzik, T. Infliximab induces apoptosis in monocytes from patients with chronic active Crohn’s disease by using a caspase-dependent pathway. Gastroenterology 2001, 121, 1145–1157. [Google Scholar] [CrossRef] [PubMed]
- Skender, B.; Hofmanova, J.; Slavik, J.; Jelinkova, I.; Machala, M.; Moyer, M.P.; Kozubik, A.; Hyrslova Vaculova, A. DHA-mediated enhancement of TRAIL-induced apoptosis in colon cancer cells is associated with engagement of mitochondria and specific alterations in sphingolipid metabolism. Biochim. Biophys. Acta 2014, 1841, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Worsham, M.J.; Lu, M.; Chen, K.M.; Stephen, J.K.; Havard, S.; Schweitzer, V.P. Malignant and nonmalignant gene signatures in squamous head and neck cancer. J. Oncol. 2012, 2012, 752860. [Google Scholar] [CrossRef] [PubMed]
- Hikita, H.; Kodama, T.; Shimizu, S.; Li, W.; Shigekawa, M.; Tanaka, S.; Hosui, A.; Miyagi, T.; Tatsumi, T.; Kanto, T.; et al. Bak deficiency inhibits liver carcinogenesis: A causal link between apoptosis and carcinogenesis. J. Hepatol. 2012, 57, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.X.; Pervaiz, S. Bcl-2 induces pro-oxidant state by engaging mitochondrial respiration in tumor cells. Cell Death Differ. 2007, 14, 1617–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Huang, Z.; Wang, Z.; Yin, C.; Zhou, L.; Zhang, L.; Huang, K.; Zhou, H.; Jiang, X.; Li, J.; et al. Identification of novel molecular markers for prognosis estimation of acute myeloid leukemia: Over-expression of PDCD7, FIS1 and Ang2 may indicate poor prognosis in pretreatment patients with acute myeloid leukemia. PLoS ONE 2014, 9, e84150. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.P.; Wang, D.; Kaushal, A.; Saligan, L. Mitochondria-Related Gene Expression Changes Are Associated with Fatigue in Patients with Nonmetastatic Prostate Cancer Receiving External Beam Radiation Therapy. Cancer Nurs. 2012, 36, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Chen, W.X.; Lv, X.B.; Tang, Q.L.; Sun, L.J.; Liu, B.D.; Zhong, J.L.; Lin, Z.Y.; Wang, Y.Y.; Li, Q.X.; et al. miR-483–5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1. Cancer Lett. 2015, 362, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Klionsky, D.J. Participation of mitochondrial fission during mitophagy. Cell Cycle Tex. 2013, 12, 3131–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nacht, M.; Ferguson, A.T.; Zhang, W.; Petroziello, J.M.; Cook, B.P.; Gao, Y.H.; Maguire, S.; Riley, D.; Coppola, G.; Landes, G.M.; et al. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999, 59, 5464–5470. [Google Scholar] [PubMed]
- Akahira, J.; Sugihashi, Y.; Suzuki, T.; Ito, K.; Niikura, H.; Moriya, T.; Nitta, M.; Okamura, H.; Inoue, S.; Sasano, H.; et al. Decreased expression of 14-3-3ε is associated with advanced disease in human epithelial ovarian cancer: Its correlation with aberrant DNA methylation. Clin. Cancer Res. 2004, 10, 2687–2693. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Gao, G.; Wang, L.; Wang, T.; Yu, J.; Zhao, Z. Stratifin expression is a novel prognostic factor in human gliomas. Path. Res. Pract. 2011, 207, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Urano, T.; Takahashi, S.; Suzuki, T.; Fujimura, T.; Fujita, M.; Kumagai, J.; Horie-Inoue, K.; Sasano, H.; Kitamura, T.; Ouchi, Y.; et al. 14-3-3ε is down-regulated in human prostate cancer. Biochem. Biophys. Res. Commun. 2004, 319, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Perathoner, A.; Pirkebner, D.; Brandacher, G.; Spizzo, G.; Stadlmann, S.; Obrist, P.; Margreiter, R.; Amberger, A. 14-3-3ε expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin. Cancer Res. 2005, 11, 3274–3279. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.Z.; Wang, J.S.; Pan, G.Q.; Lv, H.; Wen, J.F.; Luo, G.Q.; Wang, K.S.; Zhang, P.F. Comparative proteomic analysis of β-catenin-mediated malignant progression of esophageal squamous cell carcinoma. Dis. Esophagus 2010, 23, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, K.; Zhang, J.; Liu, S.S.; Dai, L.; Zhang, J.Y. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J. Proteome Res. 2011, 10, 2863–2872. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Kita, Y.; Yokomakura, N.; Uchikado, Y.; Setoyama, T.; Sakurai, H.; Omoto, I.; Matsumoto, M.; Owaki, T.; Ishigami, S.; et al. Nuclear expression of 14-3-3ε is related to prognosis in patients with esophageal squamous cell carcinoma. Anticancer Res. 2010, 30, 5175–5179. [Google Scholar] [PubMed]
- Okumura, H.; Natsugoe, S.; Matsumoto, M.; Yokomakura, N.; Uchikado, Y.; Takatori, H.; Ishigami, S.; Takao, S.; Aikou, T. Predictive value of p53 and 14-3-3ε for the effect of chemoradiation therapy on esophageal squamous cell carcinoma. J. Surg. Oncol. 2005, 91, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.Z.; Pan, G.Q.; Wang, J.S.; Wen, J.F.; Wang, K.S.; Luo, G.Q.; Shan, X.Z. Reduced stratifin expression can serve as an independent prognostic factor for poor survival in patients with esophageal squamous cell carcinoma. Dig. Dis. Sci. 2010, 55, 2552–2560. [Google Scholar] [CrossRef] [PubMed]
- Modica-Napolitano, J.S.; Aprille, J.R. Delocalized lipophilic cations selectively target the mitochondria of carcinoma cells. Adv. Drug Deliv. Rev. 2001, 49, 63–70. [Google Scholar] [CrossRef]
- Indran, I.R.; Tufo, G.; Pervaiz, S.; Brenner, C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim. Biophys. Acta 2011, 1807, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, X.; Johnson, R.H.; Kelbauskas, L.; Zhang, W.; Meldrum, D.R. Single-cell analysis reveals early manifestation of cancerous phenotype in pre-malignant esophageal cells. PLoS ONE 2013, 8, e75365. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Dabritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Modica-Napolitano, J.S.; Nalbandian, R.; Kidd, M.E.; Nalbandian, A.; Nguyen, C.C. The selective in vitro cytotoxicity of carcinoma cells by AZT is enhanced by concurrent treatment with delocalized lipophilic cations. Cancer Lett. 2003, 198, 59–68. [Google Scholar] [CrossRef]
- Mai, S.; Klinkenberg, M.; Auburger, G.; Bereiter-Hahn, J.; Jendrach, M. Decreased expression of Drp1 and Fis1 mediates mitochondrial elongation in senescent cells and enhances resistance to oxidative stress through PINK1. J. Cell Sci. 2010, 123, 917–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelissier-Rota, M.A.; Pelosi, L.; Meresse, P.; Jacquier-Sarlin, M.R. Nicotine-induced cellular stresses and autophagy in human cancer colon cells: A supportive effect on cell homeostasis via up-regulation of Cox-2 and PGE production. Int. J. Biochem. Cell Biol. 2015, 65, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, E.; Armour, S.M.; Harris, M.H.; Thompson, C.B. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003, 10, 709–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phelan, J.J.; MacCarthy, F.; O’Toole, D.; Ravi, N.; Reynolds, J.V.; O’Sullivan, J. The Mitochondrial Genes BAK1, FIS1 and SFN are Linked with Alterations in Mitochondrial Membrane Potential in Barrett’s Esophagus. Int. J. Mol. Sci. 2018, 19, 3483. https://doi.org/10.3390/ijms19113483
Phelan JJ, MacCarthy F, O’Toole D, Ravi N, Reynolds JV, O’Sullivan J. The Mitochondrial Genes BAK1, FIS1 and SFN are Linked with Alterations in Mitochondrial Membrane Potential in Barrett’s Esophagus. International Journal of Molecular Sciences. 2018; 19(11):3483. https://doi.org/10.3390/ijms19113483
Chicago/Turabian StylePhelan, James J., Finbar MacCarthy, Dermot O’Toole, Narayanasamy Ravi, John V. Reynolds, and Jacintha O’Sullivan. 2018. "The Mitochondrial Genes BAK1, FIS1 and SFN are Linked with Alterations in Mitochondrial Membrane Potential in Barrett’s Esophagus" International Journal of Molecular Sciences 19, no. 11: 3483. https://doi.org/10.3390/ijms19113483




