Detection of Plant miRNAs Abundance in Human Breast Milk
Abstract
:1. Introduction
2. Results
2.1. RNA Quality and Concentration
2.2. Plant miRNAs Are Detectable in Human Breast Milk via qRT-PCR
2.3. Relative qRT-PCR
2.4. In silico Evaluation of the Identified Plant miRNAs Influence on the Infant
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Milk Sample Collection
4.3. Total RNA Extraction from Whole Milk and Exosome Fraction
4.4. RNA Analysis and Quantifications
4.5. Reverse Transcription
4.6. Quantitative PCR Analysis
4.7. Prediction and Annotation of Putative Human Target Genes for Identified Plant miRNAs
4.8. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lessen, R.; Kavanagh, K. Position of the academy of nutrition and dietetics: Promoting and supporting breastfeeding. J. Acad. Nutr. Diet 2015, 115, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Butte, N.; Lopez-Alarcon, M.; Garza, C. Nutrient Adequacy of Exclusive Breastfeeding for the Term Infant during the First Six Months of Life. Available online: http://www.who.int/nutrition/publications/infantfeeding/9241562110/en/ (accessed on 20 May 2017).
- Laiho, K.; Lampi, A.M.; Hamalainen, M.; Moilanen, E.; Piironen, V.; Arvola, T.; Syrjanen, S.; Isolauri, E. Breast milk fatty acids, eicosanoids, and cytokines in mothers with and without allergic disease. Pediatr. Res. 2003, 53, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Shamir, R. The benefits of breast feeding. Nestle Nutr. Inst. Workshop Ser. 2016, 86, 67–76. [Google Scholar] [PubMed]
- Hanson, L.A.; Korotkova, M. The role of breastfeeding in prevention of neonatal infection. Semin. Neonatol. 2002, 7, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Ip, S.; Chung, M.; Raman, G.; Chew, P.; Magula, N.; DeVine, D.; Trikalinos, T.; Lau, J. Breastfeeding and Maternal and Infant Health Outcomes in Developed Countrie; Evidence Report/Technology Assessment; Agency for Healthcare Research and Quality: Rockville, MD, USA, 2007; pp. 1–186.
- Horta, B.L.; Loret de Mola, C.; Victora, C.G. Long-term consequences of breastfeeding on cholesterol, obesity, systolic blood pressure and type 2 diabetes: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Savino, F.; Benetti, S.; Liguori, S.A.; Sorrenti, M.; Cordero Di Montezemolo, L. Advances on human milk hormones and protection against obesity. Cell. Mol. Biol. 2013, 59, 89–98. [Google Scholar] [PubMed]
- Kalra, B.; Gupta, Y.; Kalra, S. Breast feeding: Preventive therapy for type 2 diabetes. J. Pak. Med. Assoc. 2015, 65, 1134–1136. [Google Scholar] [PubMed]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related micrornas are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Duan, R.D.; Brevaut-Malaty, V.; Gire, C.; Millet, V.; Simeoni, U.; Bernard, M.; Armand, M. Bioactive compounds in human milk and intestinal health and maturity in preterm newborn: An overview. Cell. Mol. Biol. 2013, 59, 108–131. [Google Scholar] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microrna-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Ipsaro, J.J.; Joshua-Tor, L. From guide to target: Molecular insights into eukaryotic rna-interference machinery. Nat. Struct. Mol. Biol. 2015, 22, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by micrornas: Contributions of translational repression and mrna decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Q.; Pan, X. Micrornas and their regulatory roles in animals and plants. J. Cell. Physiol. 2007, 210, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. Microrna functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Miska, E.A. How micrornas control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. Microrna functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells contain numerous mirnas that may change with milk removal and regulate multiple physiological processes. Int. J. Mol. Sci. 2016, 17, 956. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Calin, G.A. Microrna identification in plasma and serum: A new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 2009, 9, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of micrornas in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Courts, C.; Madea, B. Specific micro-rna signatures for the detection of saliva and blood in forensic body-fluid identification. J. Forensic Sci. 2011, 56, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- Korzeniewski, N.; Tosev, G.; Pahernik, S.; Hadaschik, B.; Hohenfellner, M.; Duensing, S. Identification of cell-free micrornas in the urine of patients with prostate cancer. Urol. Oncol. 2015, 33, 16.e17–16.e22. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microrna spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- De Guire, V.; Robitaille, R.; Tetreault, N.; Guerin, R.; Menard, C.; Bambace, N.; Sapieha, P. Circulating mirnas as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: Promises and challenges. Clin. Biochem. 2013, 46, 846–860. [Google Scholar] [CrossRef] [PubMed]
- Jacob, N.K.; Cooley, J.V.; Yee, T.N.; Jacob, J.; Alder, H.; Wickramasinghe, P.; Maclean, K.H.; Chakravarti, A. Identification of sensitive serum microrna biomarkers for radiation biodosimetry. PLoS ONE 2013, 8, e57603. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. Micrornas in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Modepalli, V.; Kumar, A.; Hinds, L.A.; Sharp, J.A.; Nicholas, K.R.; Lefevre, C. Differential temporal expression of milk mirna during the lactation cycle of the marsupial tammar wallaby (Macropus eugenii). BMC Genom. 2014, 15, 1012. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk mirnas primarily originate from the mammary gland resulting in unique mirna profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. Microrna as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Jiang, X.; Li, R.; Chen, M.; Song, W.; Li, X. The levels of human milk micrornas and their association with maternal weight characteristics. Eur. J. Clin. Nutr. 2016, 70, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells and lipids conserve numerous known and novel mirnas, some of which are differentially expressed during lactation. PLoS ONE 2016, 11, e0152610. [Google Scholar] [CrossRef] [PubMed]
- Floris, I.; Billard, H.; Boquien, C.Y.; Joram-Gauvard, E.; Simon, L.; Legrand, A.; Boscher, C.; Roze, J.C.; Bolanos-Jimenez, F.; Kaeffer, B. Mirna analysis by quantitative pcr in preterm human breast milk reveals daily fluctuations of hsa-mir-16-5p. PLoS ONE 2015, 10, e0140488. [Google Scholar] [CrossRef] [PubMed]
- Carney, M.C.; Tarasiuk, A.; DiAngelo, S.L.; Silveyra, P.; Podany, A.; Birch, L.L.; Paul, I.M.; Kelleher, S.; Hicks, S.D. Metabolism-related micrornas in maternal breast milk are influenced by premature delivery. Pediatr. Res. 2017, 82, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xi, Q.Y.; Ye, R.S.; Cheng, X.; Qi, Q.E.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; Jiang, Q.Y.; et al. Exploration of micrornas in porcine milk exosomes. BMC Genom. 2014, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, M.; Wang, T.; Liang, Y.; Zhong, Z.; Wang, X.; Zhou, Q.; Chen, L.; Lang, Q.; He, Z.; et al. Lactation-related microrna expression profiles of porcine breast milk exosomes. PLoS ONE 2012, 7, e43691. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microrna and mrna and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Du, X.; Li, J.; Lonnerdal, B. Human milk exosomes and their micrornas survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk disrupts p53 and dnmt1, the guardians of the genome: Implications for acne vulgaris and prostate cancer. Nutr. Metab. 2017, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Schmitz, G. Milk’s role as an epigenetic regulator in health and disease. Diseases 2017, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Park, M.R.; Son, S.J.; Kim, Y. Comparison of total rna isolation methods for analysis of immune-related micrornas in market milks. Korean J. Food Sci. Anim. Resour. 2015, 35, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microrna and messenger rna that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of micrornas in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Lee, C.H.; Laffont, B.; Savard, P.; Laugier, J.; Boilard, E.; Gilbert, C.; Fliss, I.; Provost, P. Commercial dairy cow milk micrornas resist digestion under simulated gastrointestinal tract conditions. J. Nutr. 2016, 146, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. Microrna expression profiles of bovine milk exosomes in response to staphylococcus aureus infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Chen, X.; Yu, J.; Zen, K.; Zhang, C.Y.; Li, L. Immune modulatory function of abundant immune-related micrornas in microvesicles from bovine colostrum. Protein Cell 2013, 4, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Howard, K.M.; Jati Kusuma, R.; Baier, S.R.; Friemel, T.; Markham, L.; Vanamala, J.; Zempleni, J. Loss of mirnas during processing and storage of cow’s (bos taurus) milk. J. Agric. Food Chem 2015, 63, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, B.; Pfaffl, M.W.; Dumpler, J.; von Mutius, E.; Ege, M.J. MicroRNA in native and processed cow’s milk and its implication for the farm milk effect on asthma. J. Allergy Clin. Immunol. 2016, 137, 1893–1895.e13. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhao, Z.; Sun, L.; Li, P. Fermentation results in quantitative changes in milk-derived exosomes and different effects on cell growth and survival. J. Agric. Food Chem. 2017, 65, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. Micrornas are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, hek-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant mir168a specifically targets mammalian ldlrap1: Evidence of cross-kingdom regulation by microrna. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Zielenkiewicz, P. Plant micrornas-novel players in natural medicine? Int. J. Mol. Sci. 2016, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Perge, P.; Nagy, Z.; Decmann, A.; Igaz, I.; Igaz, P. Potential relevance of micrornas in inter-species epigenetic communication, and implications for disease pathogenesis. RNA Biol. 2017, 14, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Zielenkiewicz, P. In silico identification of plant mirnas in mammalian breast milk exosomes—A small step forward? PLoS ONE 2014, 9, e99963. [Google Scholar] [CrossRef] [PubMed]
- Bagci, C.; Allmer, J. One step forward, two steps back; xeno-micrornas reported in breast milk are artifacts. PLoS ONE 2016, 11, e0145065. [Google Scholar] [CrossRef] [PubMed]
- Eldh, M.; Lotvall, J.; Malmhall, C.; Ekstrom, K. Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol. 2012, 50, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, M.; Staffolani, S.; Nocchi, L.; Neuzil, J.; Strafella, E.; Manzella, N.; Mariotti, L.; Bracci, M.; Valentino, M.; Amati, M.; et al. Clinical significance of circulating miR-126 quantification in malignant mesothelioma patients. Clin. Biochem. 2012, 45, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hotz, T.; Broadnax, L.; Yarmarkovich, M.; Elbaz-Younes, I.; Hirschi, K.D. Anomalous uptake and circulatory characteristics of the plant-based small RNA mir2911. Sci. Rep. 2016, 6, 26834. [Google Scholar] [CrossRef] [PubMed]
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome profiling of microrna by next-gen deep sequencing reveals known and novel mirna species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564. [Google Scholar] [CrossRef] [PubMed]
- Title, A.C.; Denzler, R.; Stoffel, M. Uptake and function studies of maternal milk-derived micrornas. J. Biol. Chem. 2015, 290, 23680–23691. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The c. Elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in c. Elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Emmett, P.M. Dietary patterns during complementary feeding and later outcomes. Nestle Nutr. Inst. Workshop Ser. 2016, 85, 145–154. [Google Scholar] [PubMed]
- Smithers, L.G.; Golley, R.K.; Mittinty, M.N.; Brazionis, L.; Northstone, K.; Emmett, P.; Lynch, J.W. Dietary patterns at 6, 15 and 24 months of age are associated with iq at 8 years of age. Eur. J. Epidemiol. 2012, 27, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Okubo, H.; Crozier, S.R.; Harvey, N.C.; Godfrey, K.M.; Inskip, H.M.; Cooper, C.; Robinson, S.M. Diet quality across early childhood and adiposity at 6 years: The southampton women's survey. Int. J. Obes. 2015, 39, 1456–1462. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, P.; Megraw, M.; Hatzigeorgiou, A.G. A guide through present computational approaches for the identification of mammalian microrna targets. Nat. Methods 2006, 3, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, P.; Maragkakis, M.; Papadopoulos, G.L.; Reczko, M.; Hatzigeorgiou, A.G. Lost in translation: An assessment and perspective for computational microrna target identification. Bioinformatics 2009, 25, 3049–3055. [Google Scholar] [CrossRef] [PubMed]
- van der Vuurst de Vries, A.R.; Clevers, H.; Logtenberg, T.; Meyaard, L. Leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1) is differentially expressed during human B cell differentiation and inhibits B cell receptor-mediated signaling. Eur. J. Immunol. 1999, 29, 3160–3167. [Google Scholar] [CrossRef]
- Meyaard, L.; Adema, G.J.; Chang, C.; Woollatt, E.; Sutherland, G.R.; Lanier, L.L.; Phillips, J.H. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity 1997, 7, 283–290. [Google Scholar] [CrossRef]
- Cacalano, G.; Lee, J.; Kikly, K.; Ryan, A.M.; Pitts-Meek, S.; Hultgren, B.; Wood, W.I.; Moore, M.W. Neutrophil and b cell expansion in mice that lack the murine il-8 receptor homolog. Science 1994, 265, 682–684. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Micrornas in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [PubMed]
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-throughput assessment of microrna activity and function using microrna sensor and decoy libraries. Nat. Methods 2012, 9, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.D.; Gentner, B.; Cantore, A.; Colleoni, S.; Amendola, M.; Zingale, A.; Baccarini, A.; Lazzari, G.; Galli, C.; Naldini, L. Endogenous microrna can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007, 25, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Wojcikowski, M.; Zielenkiewicz, P. Tools4mirs—One place to gather all the tools for mirna analysis. Bioinformatics 2016, 32, 2722–2724. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. Kaas: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Grossmann, S.; Vingron, M.; Robinson, P.N. Ontologizer 2.0—A multifunctional tool for go term enrichment analysis and data exploration. Bioinformatics 2008, 24, 1650–1651. [Google Scholar] [CrossRef] [PubMed]
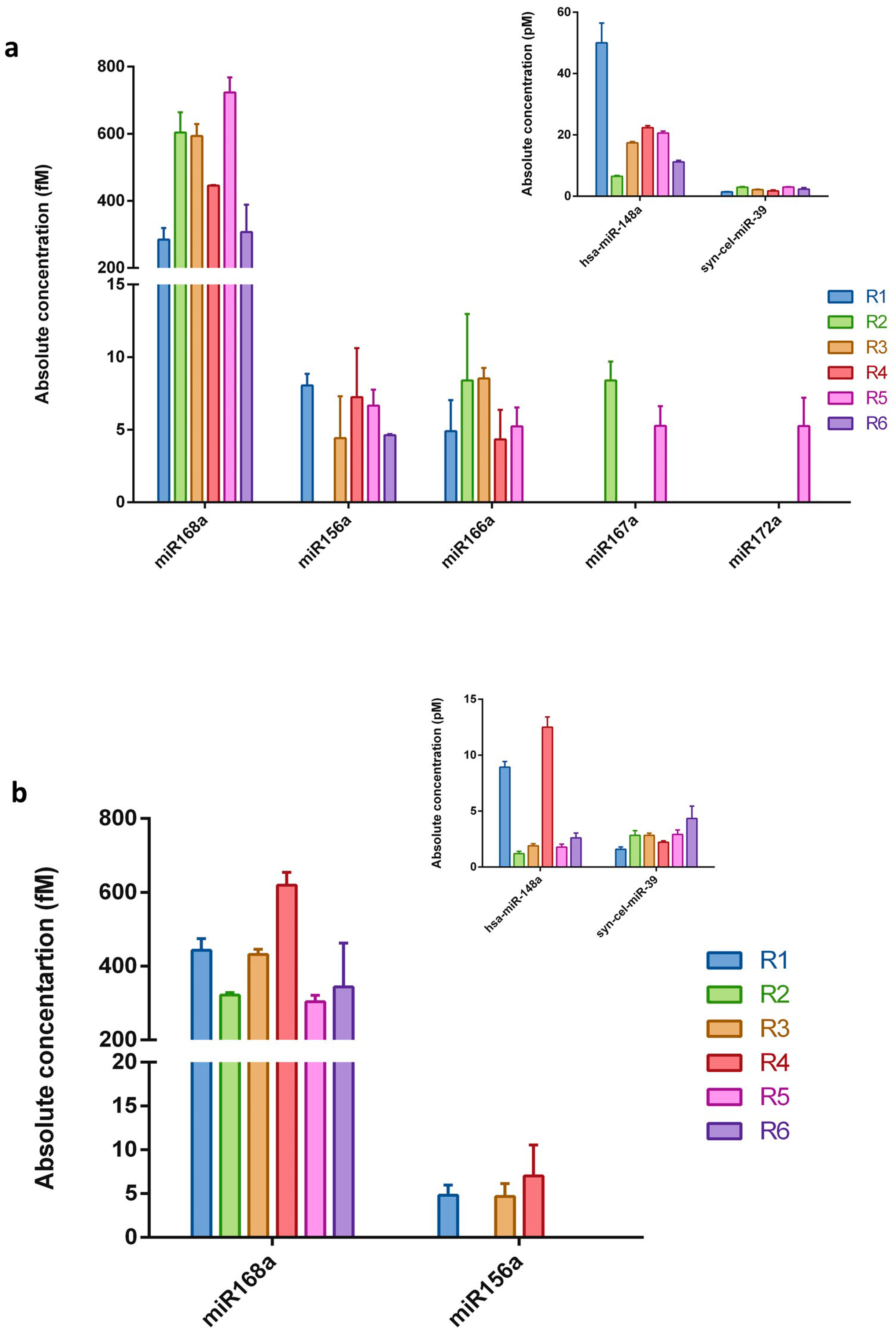
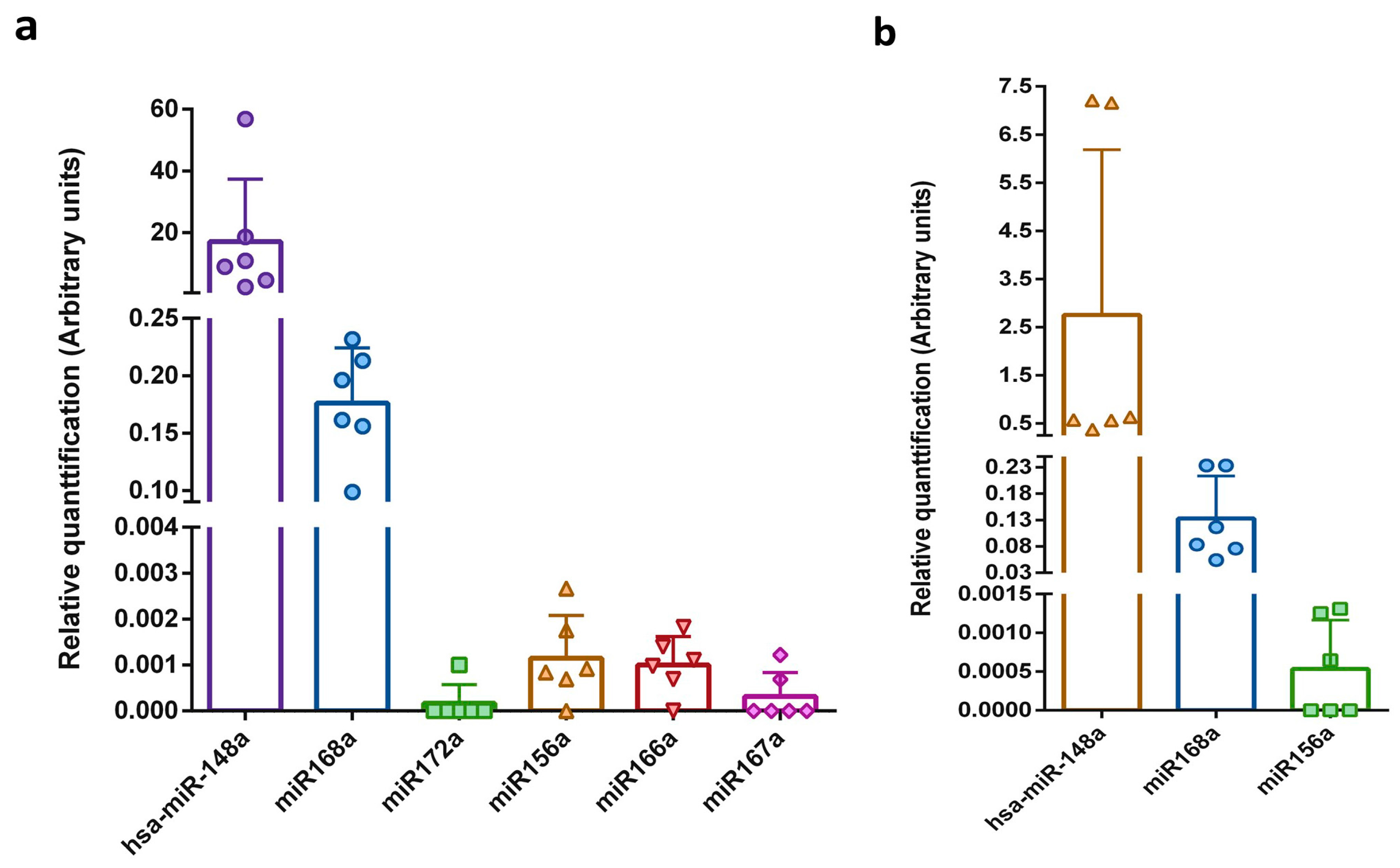
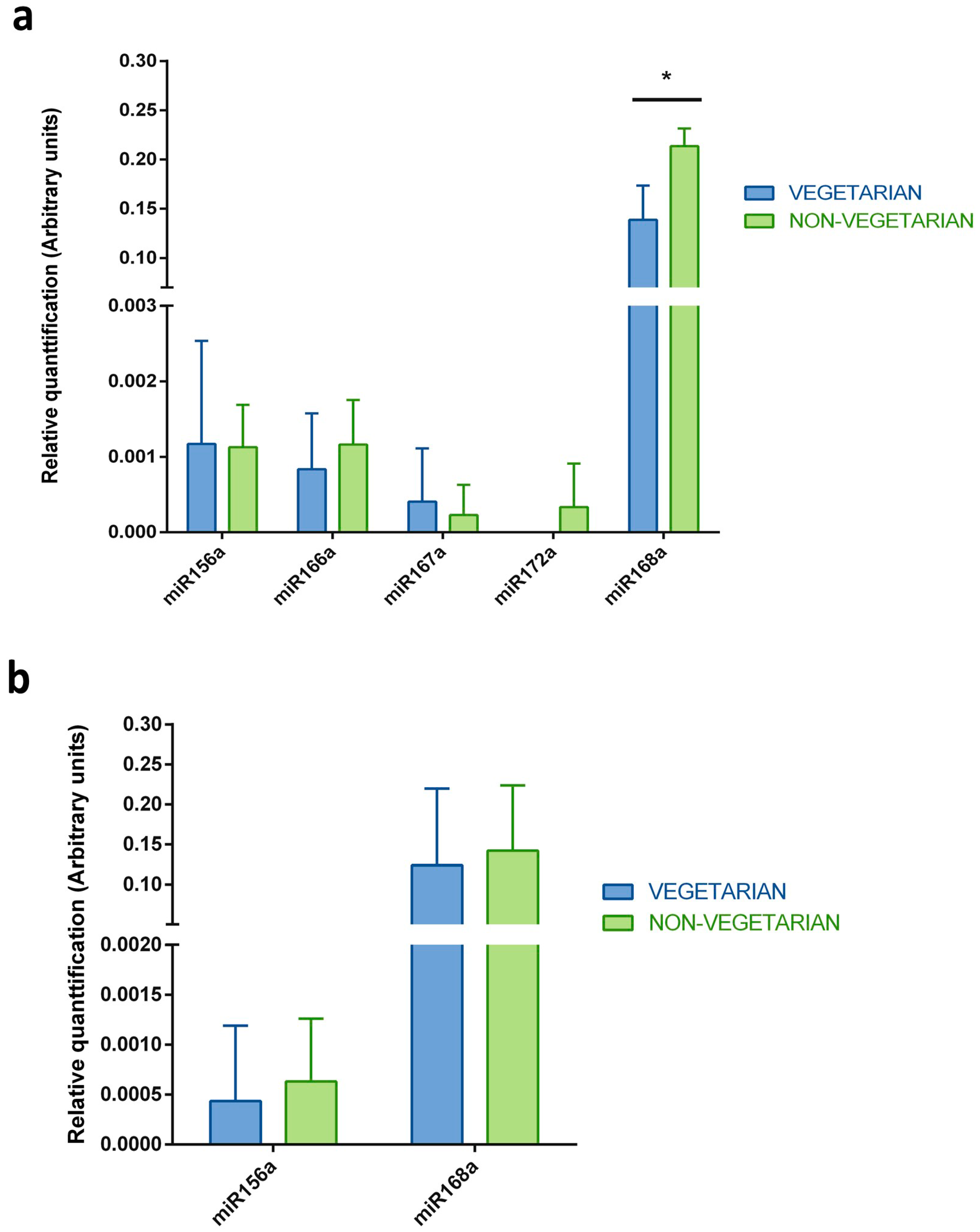
| Sample | Total RNA Concentration (ng/μL) | miRNA Concentration (ng/μL) | miRNA/Small RNA Ratio (%) | Isolation Efficiency (%) | |
|---|---|---|---|---|---|
| Whole milk | R1 | 1018 | 51.3 | 43 | 9 |
| R2 | 120 | 20.35 | 43.5 | 18.5 | |
| R3 | 94 | 15.15 | 40 | 13.4 | |
| R4 | 407 | 41 | 40.5 | 11.9 | |
| R5 | 134 | 20.25 | 43 | 18.8 | |
| R6 | 111 | 13.7 | 45 | 14.4 | |
| Exosomes | R1 | 228 | 37.19 | 19.5 | 9.95 |
| R2 | 40 | 18 | 33.5 | 17.8 | |
| R3 | 42 * | 15.45 | 29.5 | 17.7 | |
| R4 | 143 | 15.35 | 27 | 13.9 | |
| R5 | 21 * | 10.85 | 42.5 | 18.3 | |
| R6 | 23 | 9.75 | 45 | 27.15 | |
| Mothers’ ID | R6 | R1 | R2 | R4 | R5 | R3 |
|---|---|---|---|---|---|---|
| Age | 32 | 34 | 35 | 32 | 29 | 28 |
| Lactation stage (months) | 1.5 | 3.5 | 7 | 2.5 | 5 | 8 |
| Diet | vegetarian | vegetarian | vegetarian | plants included every day | plants included every day | plants included every day |
| Plants in diet up to 24 h before milk collection | tomato, pepper, cucumber, banana, pomegranate, oregano, basil, thyme, berry, barley, oat, wheat, rye | strawberries, spinach, tomato, cucumber, carrot, potato, zucchini, orange, salad, olives, mint, coriander, parsley, sesame, pumpkin seeds, sunflowers seeds, raisins, cranberry, oat, rye, millet, wheat | strawberries, tomato cucumber, carrot, salad, wheat, leek | strawberries, tomato, carrot, apple, corn, wheat, rye, kiwi | tomato, pepper, onion, cucumber, potato, radish, chive, salad, zucchini, banana, coriander, bilberry, wheat, rye | tomato, carrot, cauliflower, broccoli, plump, pear, potato, dill, rice, barley, corn, oat, rye |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukasik, A.; Brzozowska, I.; Zielenkiewicz, U.; Zielenkiewicz, P. Detection of Plant miRNAs Abundance in Human Breast Milk. Int. J. Mol. Sci. 2018, 19, 37. https://doi.org/10.3390/ijms19010037
Lukasik A, Brzozowska I, Zielenkiewicz U, Zielenkiewicz P. Detection of Plant miRNAs Abundance in Human Breast Milk. International Journal of Molecular Sciences. 2018; 19(1):37. https://doi.org/10.3390/ijms19010037
Chicago/Turabian StyleLukasik, Anna, Iwona Brzozowska, Urszula Zielenkiewicz, and Piotr Zielenkiewicz. 2018. "Detection of Plant miRNAs Abundance in Human Breast Milk" International Journal of Molecular Sciences 19, no. 1: 37. https://doi.org/10.3390/ijms19010037
APA StyleLukasik, A., Brzozowska, I., Zielenkiewicz, U., & Zielenkiewicz, P. (2018). Detection of Plant miRNAs Abundance in Human Breast Milk. International Journal of Molecular Sciences, 19(1), 37. https://doi.org/10.3390/ijms19010037





