1. Introduction
DNA topoisomerases impose changes to the topological states of circular DNA and play important roles in DNA replication, recombination, and repair as well as in RNA transcription [
1,
2]. Thermophiles encode a unique type of topoisomerase named reverse gyrase (TopR) that introduces positive supercoils to circular DNAs [
3]. Because reverse gyrase is the only protein that is exclusively conserved in thermophilic organisms, the enzyme has been implicated in genome stability maintenance in thermophiles [
4]. Indeed, genetic studies in
Thermococcus kodakaraensis, a hyperthermophilic euryarchaeon, show that growth of cells lacking TopR enzyme is significantly retarded at high temperatures (from 65 to 90 °C) [
5]. Further, crenarchaea encode two TopR proteins, but attempts to isolate TopR deletion mutants from
S. islandicus Rey15A was not successful [
6], suggesting an essential function for these enzymes in this crenarchaeon. Therefore, TopR enzymes must have important roles in thermophilic life.
Extremely thermophilic organisms have to deal with accelerated levels of spontaneous decomposition of DNA at high temperatures, including deamination of cytosine and base hydrolysis (usually depurination) [
7,
8,
9]. These naturally occurring DNA lesions often lead to the formation of apurinic/apyrimidinic (AP) sites, which eventually yields single strand DNA breaks (SSBs) [
7,
8,
9]. Likewise, base alkylation agents, such as methyl methanesulfonate (MMS), generate DNA lesions that can also be converted to AP sites and SSBs [
10]. If not repaired in a timely fashion, AP sites and SSBs induce collapse of DNA replication, giving rise to double strand DNA breaks (DSBs). DSBs are a more severe type of DNA lesion that must be repaired by homologous recombination repair (HRR), an energy-consuming pathway [
11,
12]. As a result, severe DSB damage on chromosomal DNA often leads to cell death. Therefore, one of the great challenges in thermophilic life is to deal with numerous DNA lesions that result from spontaneous DNA decomposition in order to prevent the formation of DSBs.
Since DNA lesions produced by MMS treatment must be repaired in a similar fashion as DNA lesions generated from spontaneous base decomposition in DNA, MMS treatment provides a useful means for investigating DNA damage repair mechanisms in thermophilic organisms. Indeed, investigation on the effect of MMS on genome integrity maintenance in
Sulfolobus solfataricus has revealed that TopR1 is subjected to degradation upon a lethal MMS treatment and that MMS-induced TopR1 degradation coincides with genomic DNA degradation [
13]. Moreover, it has been shown that the
Archaeoglobus fulgidus TopR binds to DNA nicks in vitro and inhibits heat-induced degradation of nicked DNA, and the DNA protection mechanism is independent of supercoiling [
14]. This raises an interesting question as to whether TopR proteins could protect damaged DNA from thermo-degradation in vivo.
In the article, we employed
S. islandicus, a genetic model for which very versatile genetic tools have been developed [
15], to investigate the function of TopR1 in genome integrity maintenance. We found that MMS treatment induced a series of events including: (a) massive breaks on the sugar-phosphate backbone of genomic DNA, (b) gradual DNA content reduction and TopR1 degradation, and (c) DNA-less cell formation and chromatin protein degradation. Moreover, we constructed a TopR1 knockdown strain (pKD-topR1/E233S1) with a CRISPR-based RNA interference tool (CRISPRi) [
16]. The strain exhibits a higher sensitivity to MMS and an early onset of DNA degradation during MMS treatment, indicating that TopR1 protects DNA breaks from thermo-degradation in vivo.
3. Discussion
In this study, we analyzed the effects of MMS on the genome integrity of S. islandicus, a thermophilic crenarchaeon and investigated the functions of TopR1 in genome stability maintenance. Several lines of experimental evidence indicate that SisTopR1 protects MMS-induced DNA breaks from thermo-degradation in a chromosome-associated manner in vivo.
First, we have demonstrated that MMS treatment immediately induces genomic DNA breakage in
Sulfolobus. MMS does not directly introduce DNA breaks to genomic DNA in yeast and mammalian cells; however, MMS-induced DNA lesions could be converted into DNA breaks by base excision repair (BER) processing or DNA replication collapse [
10]. In addition, MMS-induced DNA lesions are heat-labile in vitro and are converted to single stranded DNA breaks (SSBs) at 50 °C; two close SSBs will results in the formation of a DSB [
18]. Therefore, the DNA breaks observed in this study can be derived from two main sources: (i) thermo-degradation of MMS-induced lesions at the physiologic growth temperature of
S. islandicus and (ii) processing of the lesions by BER pathway. Considering the rapid occurrence of genomic DNA breakage, it is more likely that thermo-degradation of MMS-induced lesions constitutes the main source of MMS-induced DNA breaks. As a result, this suggests that MMS can function as a direct DNA break agent in thermophiles.
Second, lethal MMS-induced genomic DNA degradation is evidenced by the decline of DNA fragments as shown by PFGE analysis and gradual reduction of cellular DNA content as shown in flow cytometry analysis. The degradation could be directly triggered by MMS-induced massive SSBs, since an SSB can accelerate thermo-degradation of genomic DNA in vitro: first, supercoiled DNA is not likely to be denatured, while nicks initiate dsDNA denaturation [
7]; second, the exposed ssDNA is even more fragile, exhibiting a much higher depurination rate and breakage rate, leading to dsDNA breakage and further degradation [
8,
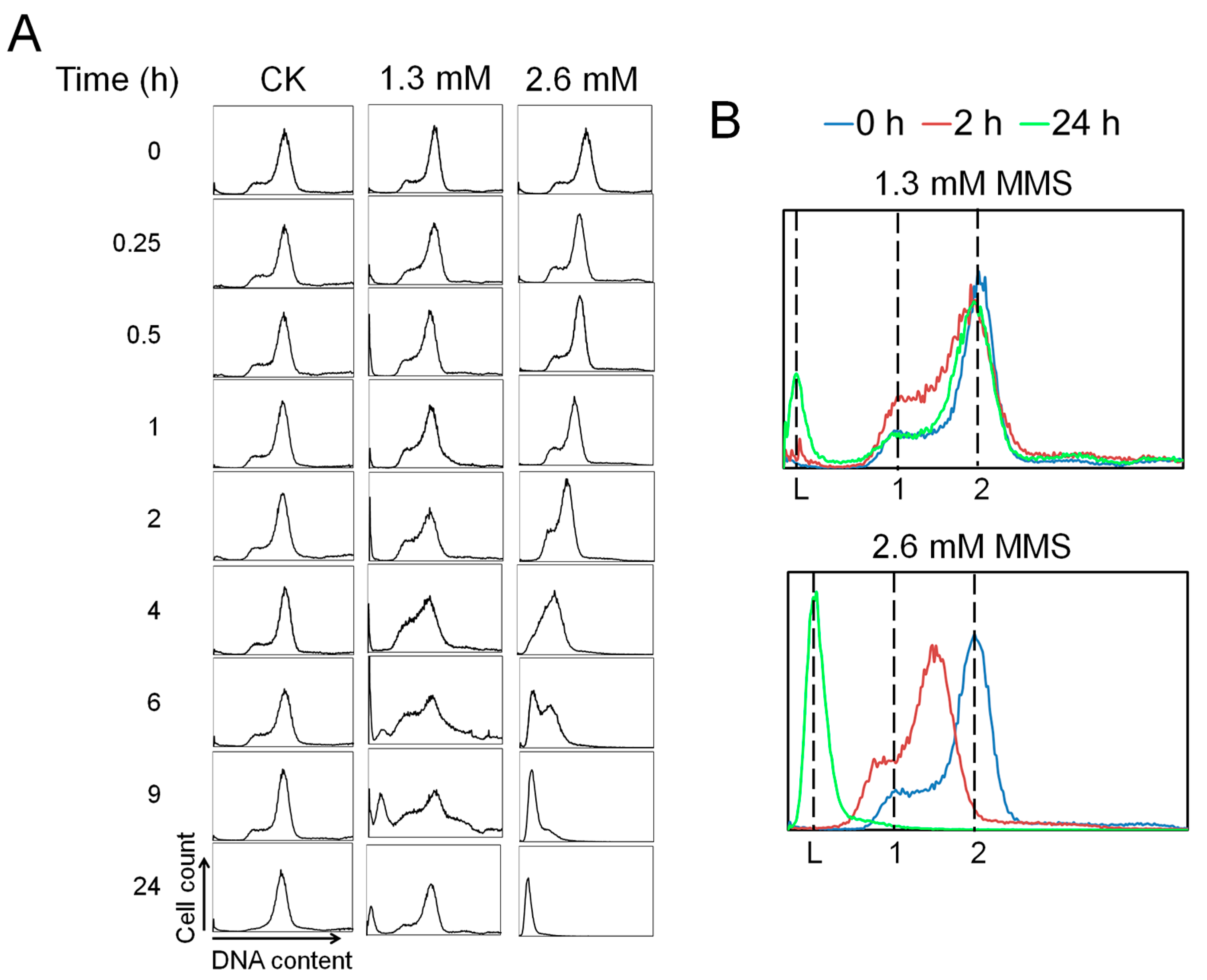
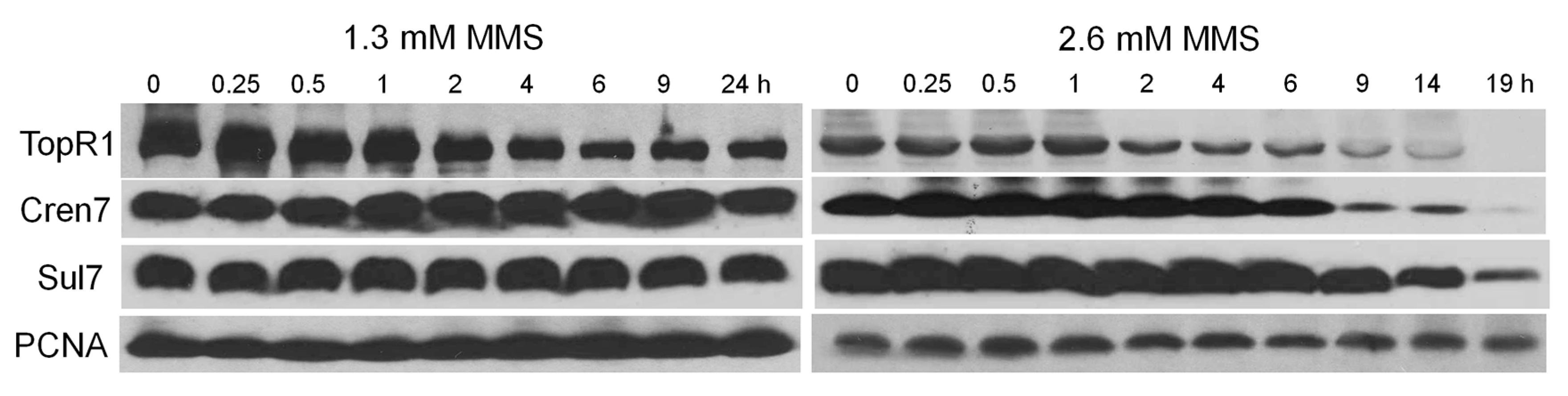
9]. More strikingly, we demonstrate that even though MMS induces immediate genomic DNA backbone breakage, cellular DNA content does not start to reduce until TopR1 degradation occurs (
Figure 2,
Figure 3 and
Figure 4), supporting the previous hypothesis that TopR functions in protecting damaged DNA from degradation [
13,
14]. Further, chromatin protein degradation occurs after TopR1 degradation and coincides with DNA-less cell formation, a common cell death consequence of the treatment of different DNA damage agents (submitted). The sequential degradation of TopR1 and chromatin proteins suggest that after MMS introduces DNA breaks, TopR1 is first subjected to degradation, leading to genomic DNA degradation, which triggers a cell death signal and induces chromatin protein degradation and DNA-less cell formation. The findings also suggest that TopR1 and chromatin proteins play distinct roles in protecting genomic DNA. We reason that TopR1 might bind to the regions that are loosely compacted or not compacted by chromatin proteins. Nevertheless, the detailed mechanisms for MMS-induced DNA degradation and the roles of these chromatin architecture proteins remain to be clarified.
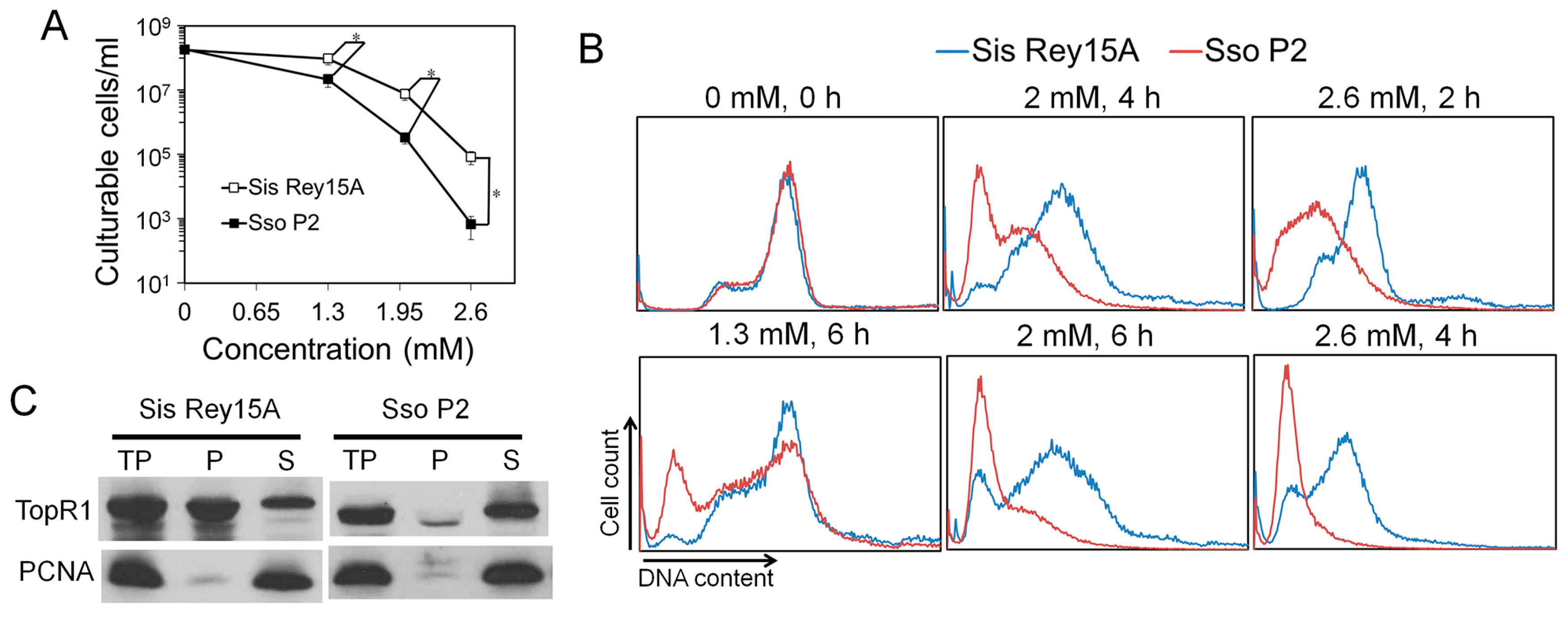
Lastly, the roles of TopR1 in genome stability maintenance have been demonstrated by CRISPRi of TopR1 expression. The strain with decreased TopR1 levels (pKD-topR1/E233S1) is more sensitive to MMS treatment than the reference strain (pSeSD1/E233S1). Further, MMS-induced DNA degradation is strongly accelerated in pKD-topR1/E233S1, suggesting that damaged DNA is prone to thermos-degradation upon an insufficiency of TopR1. The data, together with the fact that MMS induces SSBs at high temperature in vitro [
18] and rapid genomic DNA breakage in
Sulfolobus (
Figure 2), indicate that TopR1 functions in the thermo-protection of SSBs in vivo. Conceivably, TopR1 should also protect the DNA breaks derived from spontaneous DNA hydrolysis at high temperatures, which could represent a general function for reverse gyrase in thermophiles.
In addition, knockdown of TopR1 by CRISPRi in
S. islandicus cells also leads to a moderate accumulation of S phase cells. This either suggests that (1) TopR1 has a role in DNA replication or that (2) the reduced level of protection for spontaneous genomic DNA breaks in TopR1-CRISPRi cells yields DNA replication fork collapse. Indeed, it has been suggested that TopR1 is involved in cell duplication in
S. solfataricus [
19]. On the other hand, we observed that moderate MMS treatment also induces accumulation of S phase cells (
Figure 3), supporting that unrepaired DNA breaks arrest DNA replication in TopR1-CRISPRi cells. Currently, it is not possible to distinguish the two possible mechanisms experimentally.
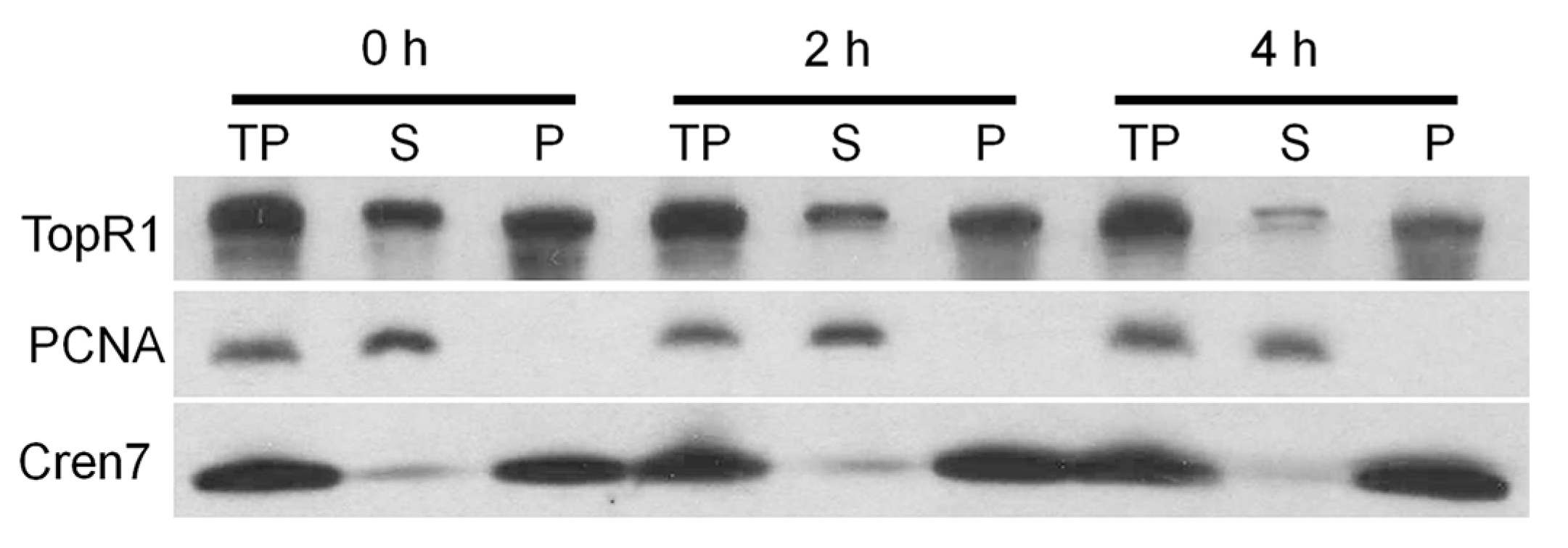
Moreover, the association of SisTopR1 to chromatin and the higher MMS resistance of
S. islandicus suggest that the DNA protection role of TopR1 is related to its association to DNA. The hypothesis is consistent with the fact that AfuTopR protects nicked DNA possibly by binding to the breaks in vitro [
14]. On the other hand, even though TopR1 proteins from
S. islandicus and
S. solfataricus exhibit high sequence similarity (
Figure S2), most SsoTopR1 protein exists in the cytoplasm fraction. Further, SsoTopR1 is recruited to the chromatin-rich fraction upon ultraviolet (UV) treatment, suggesting translocation of TopR1 upon DNA damage [
17]. A similar translocation mechanism might be adopted in
S. islandicus under normal growth conditions.
TopR1 degradation has been proved to be very specific. It was only observed during MMS treatment, while other stressful conditions, including hydroxyurea (HU) and UV treatment, fail to induce the phenomena [
13], even though UV also induces DNA breaks and arrests DNA replication in
S. solfataricus [
20]. We reason that MMS and UV induce different types of DNA breaks: MMS-induced heat-labile DNA lesions are immediately converted to massive SSBs at the physiologic growth temperature of
Sulfolobus, while UV-induced DNA lesions may be converted to DNA breaks by DNA replication fork collapse. The findings hint that TopR1 is specifically induced by massive SSBs.
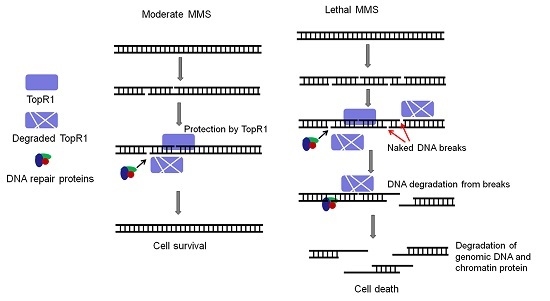
Nevertheless, the question regarding the biological meaning of TopR1 degradation remains to be answered. Possibly, TopR1 degradation represents an important step in the archetypal programmed cell death, as hypothesized previously [
13], such that TopR1 degradation results in thermo-degradation of damaged DNA, leading to cell death. However, we have shown that TopR1 is degraded to a similar extent from 2 to 6 h in the moderate and the lethal MMS treatment (
Figure 4 and
Figure 5A). The difference between the treatments is that TopR1 level recovered from 6 to 24 h during moderate MMS treatment but continuously decreased upon lethal MMS treatment. Therefore, we assume that TopR1 degradation also plays a role in DNA repair to increase the local access to DNA repair machineries of the damaged site upon moderate treatment similar to that in Eukaryotes [
21,
22]. Indeed, TopR-coated dsDNA exhibits limited accessibility in vitro [
14]. Furthermore, the two hypotheses are not mutually exclusive: upon moderate MMS treatment, TopR1 degradation increases the local access to DNA repair machineries and facilitates DNA repair and cell survival; upon lethal MMS treatment, DNA repair machineries may fail to repair massive DNA breaks in a timely fashion. As a result, the decrease of cellular TopR1 content would result in exposure of more DNA backbone breaks from thermo-degradation, leading to cell death.
4. Materials and Methods
4.1. Construction of the TopR1 Knockdown Strain
The strategy for construction of the TopR1 (gene locus: SiRe_1581) knockdown plasmid was similar to that described in [
16]. Specifically, we chose two unique sequence regions preceded by a GAAAG motif at the 5′ end from the antisense strand as spacers to abolish DNA cleavage activity [
23]. The oligonucleotides for them are shown in the
Supplementary Table S1. The spacer fragments were prepared by annealing and extension of each pair of oligonucleotides. Then, the two spacer fragments were mixed and the mixture was used as a template for further amplification with S1-forward and S2-rev-SalI as primers. The obtained fragments were inserted into pSeSD1, an expression vector widely used in
S. islandicus [
24], between StuI and SalI. Then, the generated plasmid was transferred into
Escherichia coli DH5α, and the colonies were screened by sequencing. At last, a plasmid containing one copy of each spacer was transferred into
S. islandicus E233S1 and three transformants (pKD-topR1/E233S1) were picked and grown for further analysis. In the meantime, pSeSD1 was also transferred into the genetic host as control (pSeSD1/E233S1).
4.2. Culture Growth and Treatment
The genetic host,
S. islandicus E233S1, was grown in SCVU medium, containing 0.2% sucrose (S), 0.2% Casamino acids (C), 5 µL/mL of vitamin mixture solution (V) and 20 µg/mL uracil (U), as describe previously [
25], while other stains, including pSeSD1/E233S1, pKD-TopR1/E233S1,
S. islandicus Rey15A, and
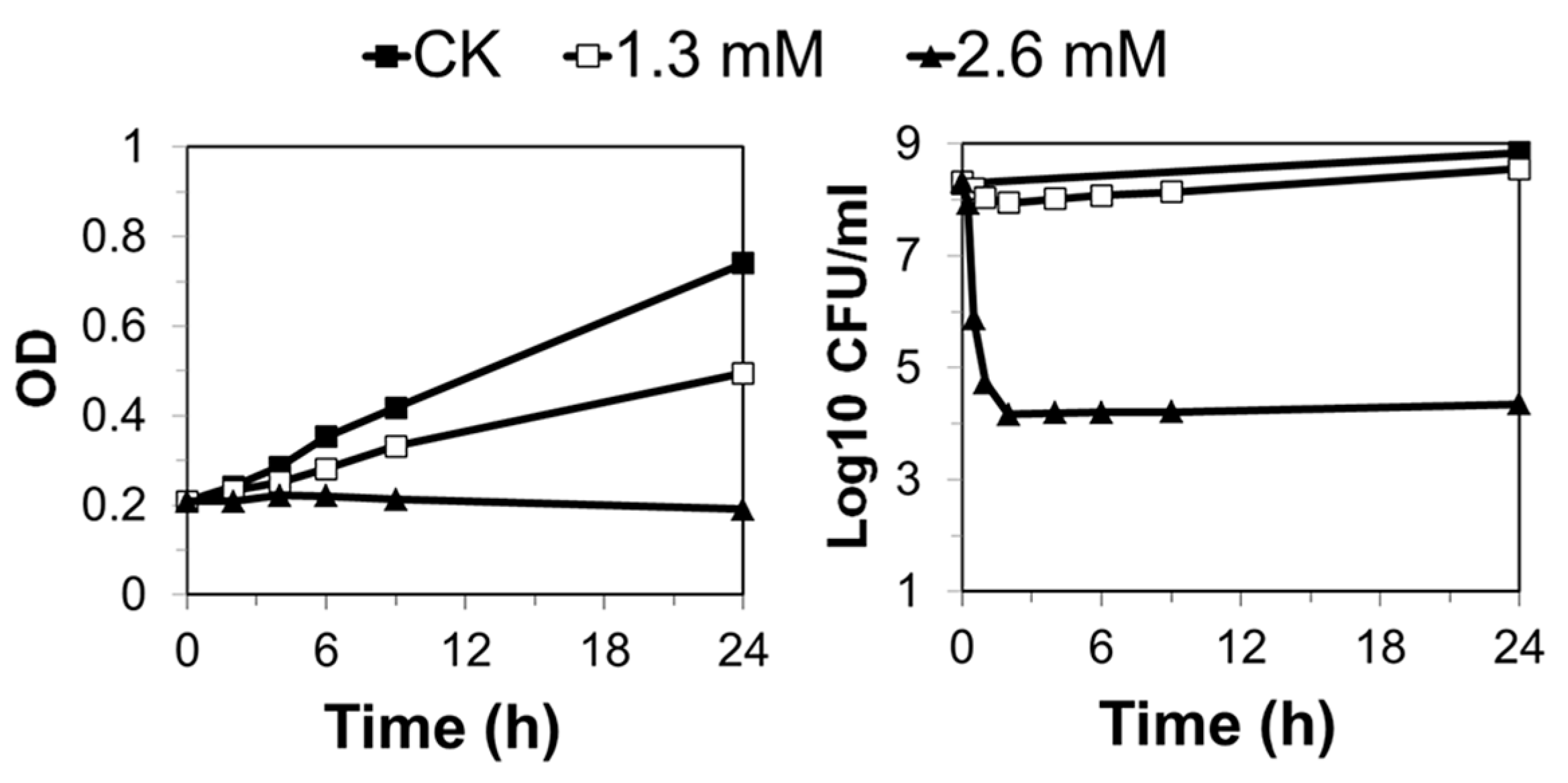
S. solfataricus P2 were grown in SCV medium that lacks uracil compared to SCVU. Before treatment, the cultures were grown at exponential phase for at least 72 h, and then early exponential phase cultures (The optical density at a wavelength of 600 nm is about 0.2) were supplemented with MMS up to the indicated concentrations. Aliquots without the drug were set as controls. During treatment, the cultures were grown at 78 °C in the dark and the samples for OD measuring, cell viability assays, flow cytometry, PFGE, and western blot were taken at indicated time points.
4.3. Cell Viability
Cells were collected from 1 mL of untreated or treated cultures by centrifugation and resuspended in 1 mL of fresh medium of the same composition. Then, resuspended cells were diluted by 10
2 and 10
4 times, and for each dilution as well as the original cell suspension, 10 µL and 100 µL of cell suspension were mixed with pre-warmed top layer gel and spread onto SCV plates as previously described (SCVU for
S. islandicus E233S1) [
25]. The plates were incubated at 75 °C for 6 days; then, plates with suitable colony densities were selected for colony counting. The cell viability was determined in three independent experiments.
4.4. Flow Cytometry
To analyze DNA content and cell size distributions, 300 µL of samples was taken from a culture to which 700 µL of absolute ethanol was added. The samples were stored at 4 °C for at least 12 h to fix cells. Then, fixed cells were collected by centrifugation at 2800 rpm for 20 min and washed with 1 mL of 10 mM Tris-NaCl, pH 7.5, and 10 mM MgCl2. Cells were collected again and resuspended in 140 µL of staining dye containing 40 µg/mL ethidium bromide (Sigma-Aldrich, St. Louis, MO, USA) and 100 µg/mL mithramycin A (Apollo chemical, Tamworth, UK). Stained cell samples were analyzed by an Apogee A40 cytometer (Apogeeflow, Hertfordshire, UK) equipped with 405 nm laser. A dataset of at least 60,000 cells was collected for each sample.
4.5. Western Blot
Cells from 10 mL of the untreated or treated cultures were collected and resuspended in bufferA (50 mM Tris-NaCl, pH 8.0, 100 mM NaCl). Then, 5× SDS-loading buffer was added into the solution, followed by heating at 95 °C for 10 min, to disrupt the cells. The final volume of the samples was calculated according to the optical density to make sure that the number of cells for each line was same. Then, the proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA) and the membrane was incubated with antibodies for Cren7 [
26], Sul7d [
27], TopR1, and PCNA [
28]. Afterwards, the membrane was incubated with anti-rabbit horseradish peroxidase (HRP) Secondary antibody (Thermo Fisher Scientific, Waltham, MA, USA) and the enhanced chemiluminescence (ECL) western blot substrate (Thermo Fisher Scientific, Waltham, MA, USA) was used to detect the signals. The antiserum against TopR1 was raised in rabbits using purified recombinant TopR1 protein.
4.6. Pulsed-Field Gel Electrophoresis (PFGE)
About 6 × 109 cells (given that 1 mL culture of OD600 = 1.0 contains 109 cells) were collected from 30 mL of untreated or treated S. islandicus culture by centrifugation. The cells were washed with 50 mL of TEN buffer (50 mM Tris-Cl, pH 8, 50 mM EDTA, pH 8, 100 mM NaCl) and resuspended in 150 µL prewarmed TEN buffer (37 °C). Then, the cell suspension was mixed with 150 µL of 0.8% low-melting-point agarose (precooled to 37 °C) and transferred to a plug mold with 85 µL per well. After solidification at 4 °C for 30 min, the plugs were released from the mold into a 1.5 mL microfuge tube containing 1 mL of NDS solution (0.5 M EDTA, 0.12% Tris, 0.55 M NaOH, pH 8.5, 1% N-lauroyl sarcosine sodium salt) and 1 mg/mL proteinase K (Sigma-Aldrich, St. Louis, MO, USA) and incubated for two days at 30 °C. The old proteinase K-containing NDS buffer was replaced by a fresh solution of the same composition after 1 day’s incubation. The plugs then were washed three times with NDS and stored at 4 °C. For PFGE, about one fourth of a plug was transferred into each well of an agarose gel, and electrophoresis was conducted with a CHEF-DR III pulsed-field electrophoresis system (Bio-Rad, Hercules, CA, USA) under the running condition of 3.5 V/cm with the switch time from 5 to 100 s for 40 h. Then, the gel was stained with a nucleic staining dye SYBR gold (Thermo Fisher Scientific, Waltham, MA, USA), and the image was captured by using Typhoon FLA 7000 (GE Healthcare, Chicago, IL, USA).
4.7. Isolation of DNA-Rich Fraction
The procedure to isolate DNA-rich fraction from
Sulfolobus was modified from that described in [
29]. Specifically, about 8 × 10
9 cells from different
Sulfolobus species were harvested by centrifugation, washed with extraction buffer (50 mM Tris-HCl pH 7, 15 mM MgCl
2, 50 mM NaCl, 1 mM DTT, 0.4 M sorbitol) and resuspended with 400 µL of the same buffer. Then, half volume of the cell solution was mixed with 5× SDS-loading buffer (total protein, TP), while the rest was supplemented with 0.5% Triton X-100, placed on ice for 30 min, and subjected to centrifugation at 13,000 rpm for 10 min at 4 °C. The supernatant (S, cytoplasm fraction) derived from the centrifugation was then mixed with 5× SDS-loading buffer, while the pellet (P, DNA-rich fraction) was resuspended with 200 µL of extraction buffer and also mixed with 5× SDS-loading buffer. At last, all three fractions were heated at 95 °C for 10 min and 10 µL of each fraction were run on SDS-gels and analyzed by western blot.