Mycoviruses in the Plant Pathogen Ustilaginoidea virens Are Not Correlated with the Genetic Backgrounds of Its Hosts
Abstract
:1. Introduction
2. Results
2.1. dsRNA Elements Were Prevalent in Most of the Screened U. virens Strains
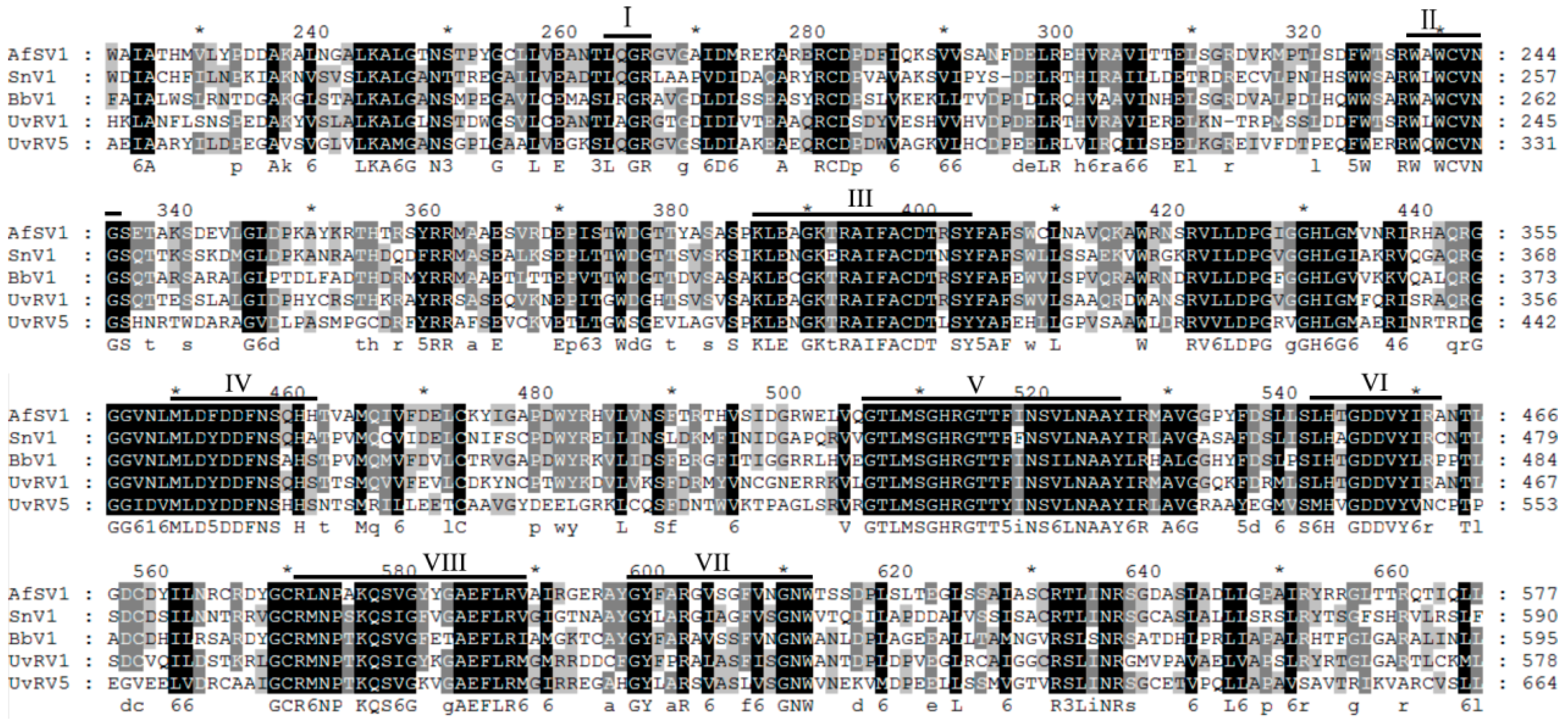
2.2. Sequence Analysis of a Novel dsRNA Mycovirus in Strain F10-338
2.3. Phylogenetic Analysis of UvRV5
2.4. Phylogenetic Analysis of IGS Sequences
2.5. dsRNA Infection is Not Genetically Related to the Host Fungi
2.6. Detection of UvRV5 in Other U. virens Strains
3. Discussion
4. Materials and Methods
4.1. Fungal Isolates and Growth Conditions
4.2. dsRNA Extraction
4.3. cDNA Synthesis, Cloning, Sequencing, and Phylogenetic Analysis
4.4. DNA Isolation
4.5. IGS Amplification, Sequencing, and Phylogenetic Analysis
4.6. Detection of UvRV5 in Other U. virens Strains
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cooke, M.C. Some extra-European fungi. Grevillea 1878, 7, 3. [Google Scholar]
- Ladhalakshmi, D.; Laha, G.; Singh, R.; Karthikeyan, A.; Mangrauthia, S.; Sundaram, R.; Thukkaiyannan, P.; Viraktamath, B. Isolation and characterization of Ustilaginoidea virens and survey of false smut disease of rice in India. Phytoparasitica 2012, 40, 171–176. [Google Scholar] [CrossRef]
- Tang, Y.X.; Jin, J.; Hu, D.W.; Yong, M.L.; Xu, Y.; He, L.P. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 2013, 62, 1–8. [Google Scholar] [CrossRef]
- Ashizawa, T.; Takahashi, M.; Arai, M.; Arie, T. Rice false smut pathogen, Ustilaginoidea virens, invades through small gap at the apex of a rice spikelet before heading. J. Gen. Virol. 2012, 78, 255–259. [Google Scholar] [CrossRef]
- Luduena, R.F.; Roach, M.C.; Prasad, V.; Banerjee, M.; Koiso, Y.; Li, Y.; Iwasaki, S. Interaction of ustiloxin A with bovine brain tubulin. Biochem. Pharmacol. 1994, 47, 1593–1599. [Google Scholar] [CrossRef]
- Koiso, Y.; Li, Y.; Iwasaki, S.; Hanaoka, K.; Kobayashi, T.; Sonoda, R.; Fujita, Y.; Yaegashi, H.; Sato, Z. Ustiloxin, antimitotic cyclic peptides from false smut balls on rice panicles caused by Ustilaginoidea virens. J. Antibiot. 1994, 47, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Fu, Y.P.; Jiang, D.H.; Ghabrial, S.A.; Li, G.Q.; Peng, Y.L.; Xie, J.T.; Cheng, J.S.; Huang, J.B.; et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, J.; Cheng, J.; Fu, Y.; Li, G.; Yi, X.; Jiang, D. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. USA 2014, 111, 12205–12210. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Jin, F.; Zhang, J.; Yang, L.; Jiang, D.; Li, G. Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J. Virol. 2012, 86, 6605–6619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, S.; Chiba, S.; Kondo, H.; Kanematsu, S.; Suzuki, N. A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front. Microbiol. 2014, 5, 360. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Zhang, H.; Qiu, D.; Guo, L. Two novel relative double-stranded RNA mycoviruses infecting Fusarium poae strain SX63. Int. J. Mol. Sci. 2016, 17, 641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jiang, Y.; Huang, J.; Dong, W. Genomic organization of a novel partitivirus from the phytopathogenic fungus Ustilaginoidea virens. Arch. Virol. 2013, 158, 2415–2419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jiang, Y.; Huang, J.; Dong, W. Complete genome sequence of a putative novel victorivirus from Ustilaginoidea virens. Arch. Virol. 2013, 158, 1403–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Zhu, J.Z.; Lei, X.H.; Chen, D.; Zhu, H.J.; Gao, B.D. Complete genome sequence and organization of a novel virus from the rice false smut fungus Ustilaginoidea virens. Virus Genes 2014, 48, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Zhou, Q.; Lei, X.H.; Chen, D.; Shang, H.H.; Zhu, H.J. The nucleotide sequence and genome organization of two victoriviruses from the rice false smut fungus Ustilaginoidea virens. Virus Genes 2014, 48, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Lei, X.H.; Zhu, J.Z.; Song, G.; Zhang, Y.D.; Chen, Y.; Gao, B.D. Detection and sequence analysis of two novel co-infecting double-strand RNA mycoviruses in Ustilaginoidea virens. Arch. Virol. 2014, 159, 3063–3070. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.J.; Chen, D.; Zhong, J.; Zhang, S.Y.; Gao, B.D. A novel mycovirus identified from the rice false smut fungus Ustilaginoidea virens. Virus Genes 2015, 51, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Luo, C.; Jiang, D.; Li, G.; Huang, J. The complete genomic sequence of a second novel partitivirus infecting Ustilaginoidea virens. Arch. Virol. 2014, 159, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jiang, Y.; Dong, W. A novel monopartite dsRNA virus isolated from the phytopathogenic fungus Ustilaginoidea virens and ancestrally related to a mitochondria-associated dsRNA in the green alga Bryopsis. Virology 2014, 462, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Nibert, M.L. Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch. Virol. 2009, 154, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Havens, W.M.; Nibert, M.L.; Ghabrial, S.A. RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190 S and other victoriviruses (Family Totiviridae). J. Virol. 2011, 85, 7343–7352. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, T.; Luo, C.; Jiang, D.; Li, G.; Li, Q.; Huang, J. Prevalence and diversity of mycoviruses infecting the plant pathogen Ustilaginoidea virens. Virus Res 2015, 195, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, M.F.; Jamal, A.; Bignell, E.M.; Petrou, M.A.; Coutts, R.H. Incidence of dsRNA mycoviruses in a collection of Aspergillus fumigatus isolates. Mycopathologia 2012, 174, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Herrero, N.; Dueñas, E.; Quesada-Moraga, E.; Zabalgogeazcoa, I. Prevalence and diversity of viruses in the entomopathogenic fungus Beauveria bassiana. Appl. Environ. Microbiol. 2012, 78, 8523–8530. [Google Scholar] [CrossRef] [PubMed]
- Herrero, N.; Zabalgogeazcoa, I. Mycoviruses infecting the endophytic and entomopathogenic fungus Tolypocladium cylindrosporum. Virus Res. 2011, 160, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Romo, M.; Leuchtmann, A.; García, B.; Zabalgogeazcoa, I. A totivirus infecting the mutualistic fungal endophyte Epichloë festucae. Virus Res. 2007, 124, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Chen, X.B.; Punja, Z.K. Diversity, complexity and transmission of double-stranded RNA elements in Chalara elegans (synanam. Thielaviopsis basicola). Mycol. Res. 2006, 110, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Peever, T.L.; Liu, Y.C.; Milgroom, M.G. Diversity of hypoviruses and other double-stranded RNAs in Cryphonectria parasitica in North America. Phytopathology 1997, 87, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, M.; Nakamura, H.; Uetake, Y.; Matsumoto, N. Presence and distribution of double-stranded RNA elements in the white root rot fungus Rosellinia necatrix. Mycoscience 2002, 43, 21–26. [Google Scholar] [CrossRef]
- Ikeda, K.I.; Nakamura, H.; Arakawa, M.; Matsumoto, N. Diversity and vertical transmission of double-stranded RNA elements in root rot pathogens of trees, Helicobasidium mompa and Rosellinia necatrix. Mycol Res. 2004, 108, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Ihrmark, K.; Zheng, J.; Stenström, E.; Stenlid, J. Presence of double-stranded RNA in Heterobasidion annosum. Forest Pathol. 2001, 31, 387–394. [Google Scholar] [CrossRef]
- Vainio, E.J.; Hakanpää, J.; Dai, Y.C.; Hansen, E.; Korhonen, K.; Hantula, J. Species of Heterobasidion host a diverse pool of partitiviruses with global distribution and interspecies transmission. Fungal Biol. 2011, 115, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Márquez, L.M.; Redman, R.S.; Rodriguez, R.J.; Roossinck, M.J. A virus in a fungus in a plant: Three-way symbiosis required for thermal tolerance. Science 2007, 315, 513–515. [Google Scholar] [CrossRef] [PubMed]
- Asensio, N.H.; Márquez, S.S.; Zabalgogeazcoa, I. Mycovirus effect on the endophytic establishment of the entomopathogenic fungus Tolypocladium cylindrosporum in tomato and bean plants. Biol. Control 2013, 58, 225–232. [Google Scholar]
- Majer, D.; Mithen, R.; Lewis, B.G.; Vos, P.; Oliver, R.P. The use of AFLP fingerprinting for the detection of genetic variation in fungi. Mycol. Res. 1996, 100, 1107–1111. [Google Scholar] [CrossRef]
- Xu, D.H.; Wahyuni, S.; Sato, Y.; Yamaguchi, M.; Tsunematsu, H.; Ban, T. Genetic diversity and relationships of Japanese peach (Prunus persica L.) cultivars revealed by AFLP and pedigree tracing. Genet. Resour. Crop. Evol. 2006, 53, 883–889. [Google Scholar] [CrossRef]
- Susanna, P.; Elisabetta, M.; Giovanni, V. Intraspecific diversity within Diaporthe helianthi: Evidence from rDNA intergenic spacer (IGS) sequence analysis. Mycopathologia 2007, 57, 317–326. [Google Scholar]
- Hsing, M.H.; Chia, L.L.; Li, C.T.; Rur, J.H.; Kuo, L.L.; Adrian, L.; James, C.L. Characterization of the polymorphic repeat sequence within the rDNA IGS of Cannabis sativa. Forensic. Sci. Int. 2005, 152, 23–28. [Google Scholar]
- Refos, J.M.; Vonk, A.G.; Eadie, K.; Lo-Ten-Foe, J.R.; Verbrugh, H.A.; van Diepeningen, A.D.; Van de Sande, W.W. Double-stranded RNA mycovirus infection of Aspergillus fumigatus is not dependent on the genetic make-up of the host. PLoS ONE 2013, 8, e77381. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.L.; Pan, Y.J.; Xie, X.W.; Zhu, L.H.; Xu, J.L.; Wang, S.; Li, Z.K. Genetic diversity of rice false smut fungus, Ustilaginoidea virens and its pronounced differentiation of populations in North China. J. Phytopathol. 2008, 156, 559–564. [Google Scholar] [CrossRef]
- Sun, X.; Kang, S.; Zhang, Y.; Tan, X.; Yu, Y.; He, H.; Cai, L. Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS ONE 2013, 8, e76879. [Google Scholar] [CrossRef]
- O’Donnell, K.; Gueidan, C.; Sink, S.; Johnston, P.R.; Crous, P.W.; Glenn, A.; van Der Lee, T. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 2009, 46, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.J.; Dodds, J.A. Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 1979, 69, 854–858. [Google Scholar] [CrossRef]
- Potgieter, A.C.; Page, N.A.; Liebenberg, J.; Wright, I.M.; Landt, O.; van Dijk, A.A. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J. Gen. Virol. 2009, 90, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Higgins, D.G. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Fan, R.H.; Liu, B.; Li, S.H.; Zheng, D.W.; Zhang, J.Z.; Hu, D.W. Prelimi nary analysis of rDNA IGS of Ustilaginoidea virens isolates from different geographical regions in China. Acta Phytopathol. Sin. 2010, 2, 018. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Boil. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Boil. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
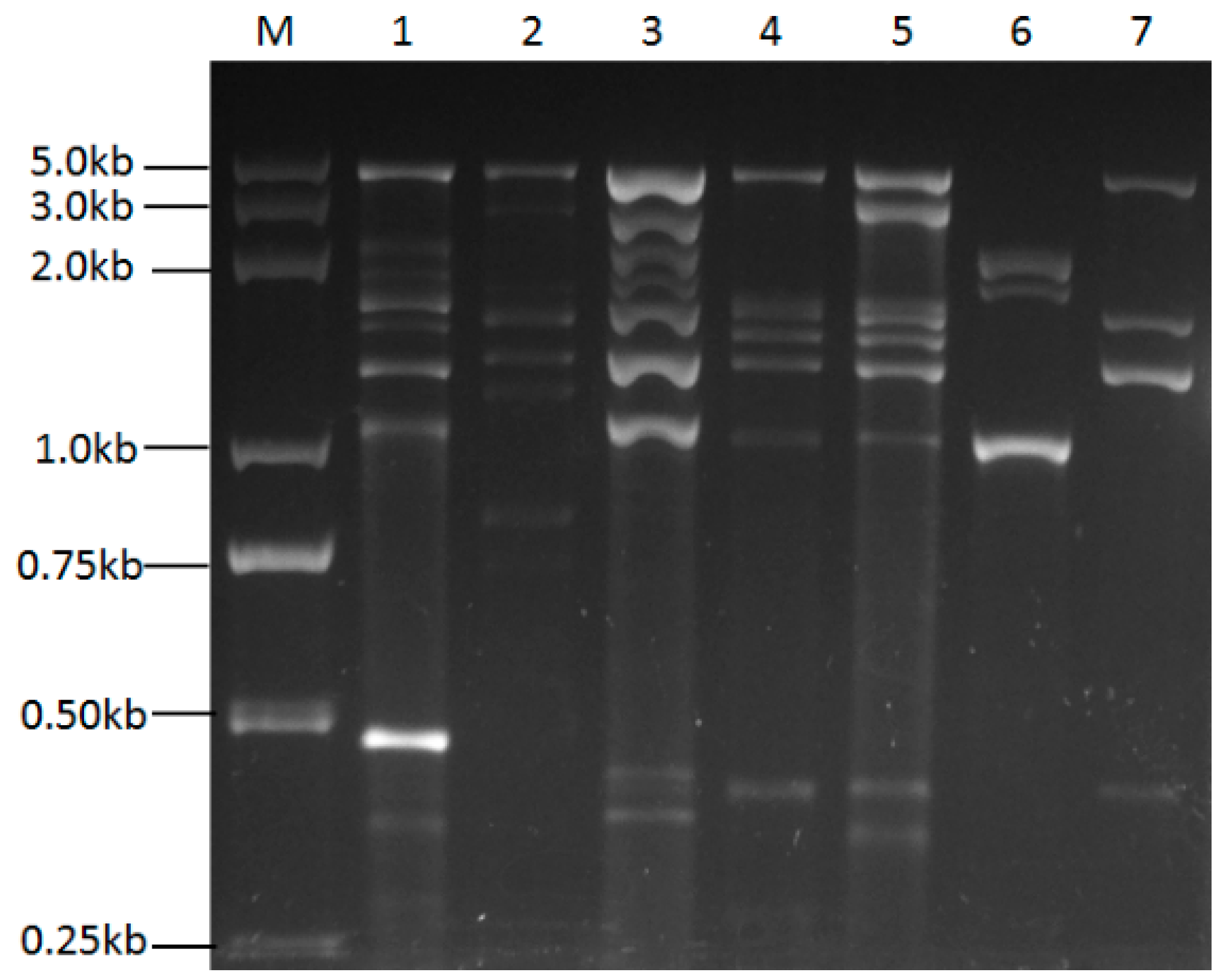
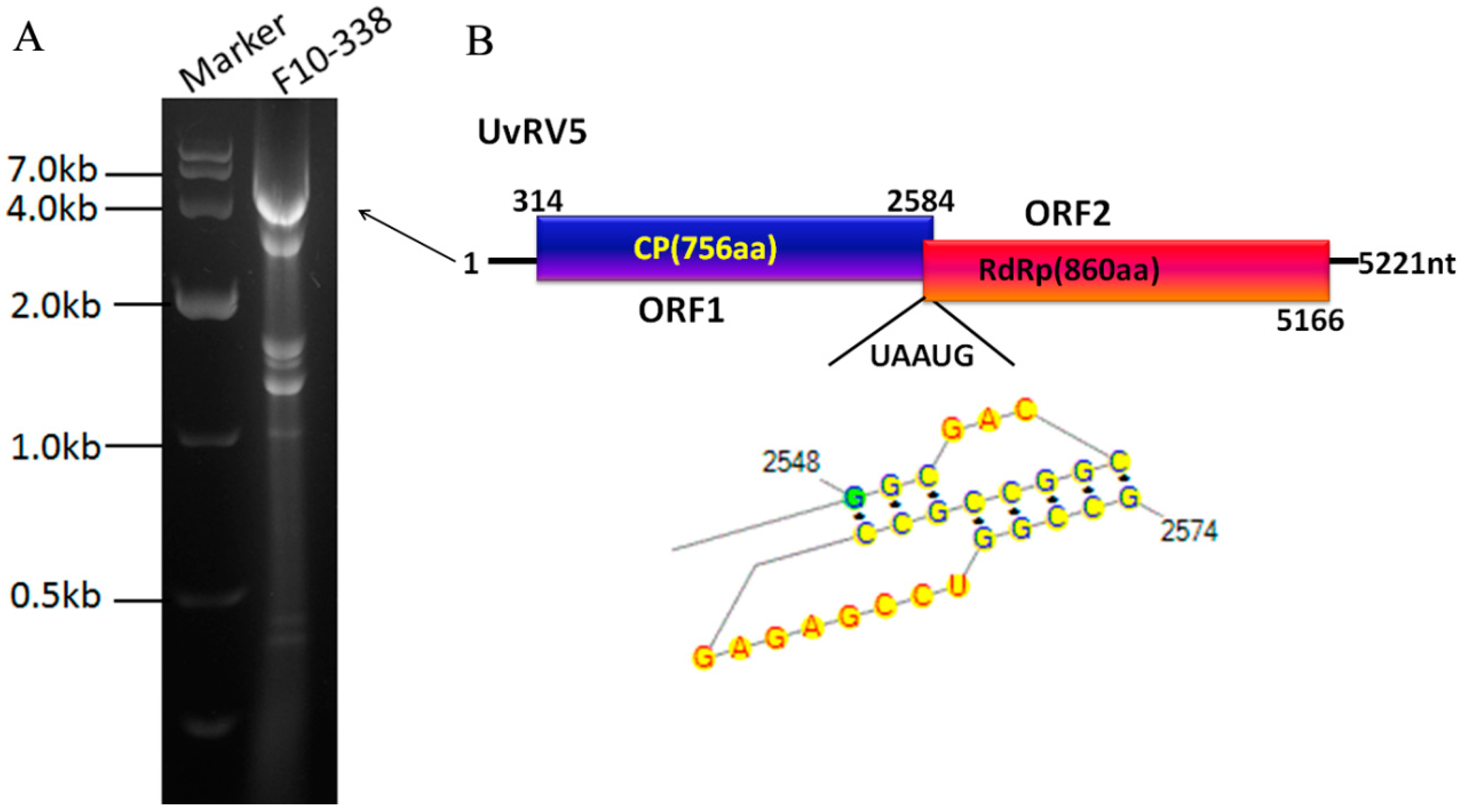




| Strains | DsRNA Elements Observed (kbp) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.3 | 0.4 | 0.5 | 0.6 | 0.75 | 1.0 | 1.2 | 1.3 | 1.4 | 1.5 | 1.6 | 2.0 | 2.5 | 2.8 | 3.0 | 4.0 | 5.3 | IGS Clade | |
| F10-54 | / | / | Clade I | |||||||||||||||
| FYD1201 | / | / | ||||||||||||||||
| F-09-3 | / | / | / | / | / | / | / | |||||||||||
| 0901 | / | / | / | |||||||||||||||
| F10-56 | / | / | / | / | / | |||||||||||||
| F10-331 | / | / | / | / | / | / | / | / | / | / | ||||||||
| F10-55 | / | / | / | / | ||||||||||||||
| F10-87 | / | |||||||||||||||||
| F10-338 | / | / | / | / | / | / | / | / | / | |||||||||
| SM03 | / | |||||||||||||||||
| FYJ1236 | / | / | / | / | / | / | / | / | ||||||||||
| F10-81 | / | / | / | / | ||||||||||||||
| F10-60 | / | / | / | |||||||||||||||
| FNH1206 | / | / | ||||||||||||||||
| F10-332 | / | / | / | |||||||||||||||
| N10-2 | / | / | / | |||||||||||||||
| F10-69 | / | / | / | |||||||||||||||
| F10-325 | / | |||||||||||||||||
| F10-15 | / | / | / | / | / | / | ||||||||||||
| F11-33 | / | / | / | |||||||||||||||
| F-09-01 | / | / | / | / | / | / | / | |||||||||||
| SM02 | / | / | / | |||||||||||||||
| F10-315 | / | / | / | Clade II | ||||||||||||||
| F10-80 | / | / | / | / | / | |||||||||||||
| F10-341 | / | / | / | / | ||||||||||||||
| Uv10 | / | / | / | / | / | / | / | / | / | / | / | / | / | / | ||||
| F10-49 | / | / | / | |||||||||||||||
| FNH1225 | / | / | / | / | / | / | / | / | / | |||||||||
| SM | / | / | / | |||||||||||||||
| Uv11 | / | / | / | / | / | / | / | / | ||||||||||
| FNH1212 | / | / | / | / | / | / | / | / | / | / | ||||||||
| F10-336 | / | / | / | / | ||||||||||||||
| FYD1202 | / | / | / | / | / | / | / | / | / | |||||||||
| NJ10-1 | / | / | / | / | / | / | / | |||||||||||
| Virus | CP | RdRp | ||||||
|---|---|---|---|---|---|---|---|---|
| Identity (%) | E Value | Overlap (Positions) | Query Coverage (%) | Identity (%) | E Value | Overlap (Positions) | Query Coverage (%) | |
| AfsV1 | 41 | 7 × 10−156 | 290/700 | 92 | 41 | 0 | 319/776 | 87 |
| SnV1 | 46 | 3 × 10−171 | 303/656 | 86 | 40 | 7 × 10−172 | 322/797 | 90 |
| BbV1 | 43 | 1 × 10−169 | 311/719 | 94 | 39 | 1 × 10−165 | 313/798 | 90 |
| UvRV1 | 43 | 6 × 10−165 | 304/714 | 94 | 40 | 1 × 10−164 | 309/780 | 89 |
| HmV1-17 | 40 | 9 × 10−148 | 255/643 | 84 | 38 | 2 × 10−163 | 299/790 | 89 |
| MoV1 | 37 | 8 × 10−111 | 227/610 | 79 | 35 | 2 × 10−147 | 280/789 | 89 |
| GaRV-L1 | 38 | 1 × 10−114 | 261/685 | 86 | 39 | 2× 10−143 | 304/786 | 89 |
| GaRV-L2 | 38 | 2 × 10−114 | 260/685 | 86 | 39 | 3 × 10−143 | 303/786 | 89 |
| MoV2 | 38 | 7 × 10−99 | 225/600 | 77 | 38 | 3 × 10−140 | 300/798 | 89 |
| PvRV | 39 | 9 × 10−113 | 245/628 | 80 | 37 | 1 × 10−138 | 289/786 | 88 |
| BfTV1 | 43 | 6 × 10−172 | 279/64 | 84 | 34 | 4 × 10−138 | 298/873 | 99 |
| TcV1 | 43 | 3 × 10−157 | 272/637 | 84 | 35 | 4 × 10−138 | 313/886 | 99 |
| UvRV3 | 37 | 1 × 10−99 | 224/606 | 78 | 37 | 9 × 10−137 | 288/784 | 88 |
| HvV190S | 46 | 3 × 10−180 | 289/632 | 83 | 36 | 2 × 10−136 | 281/788 | 89 |
| SsRV2 | 39 | 9 × 10−105 | 234/607 | 77 | 37 | 2 × 10−134 | 300/805 | 91 |
| CmRV | 38 | 4 × 10−105 | 228/602 | 78 | 37 | 5 × 10−126 | 293/783 | 88 |
| NoV1 | 36 | 9 × 10−105 | 236/655 | 84 | 36 | 8 × 10−126 | 284/786 | 88 |
| Genus | Virus Name | Abbreviation | Accession Number. | |
|---|---|---|---|---|
| CP | RdRp | |||
| Victorivirus | Ustilaginoidea virens RNA virus 5 | UvRV5 | ALP73430.1 | ALP73431.1 |
| Beauveria bassiana victorivirus 1 | BbV1 | CCC42234.1 | AMQ11131.1 | |
| Botryotinia fuckeliana totivirus 1 | BfTV1 | YP_001109579.1 | AM491608 | |
| Coniothyrium minitans RNA virus | CMRV | YP_392466.1 | AF527633 | |
| Gremmeniella abietina RNA virus L1 | GaRV-L1 | NP_624331.1 | NP_624332.2 | |
| Gremmeniella abietina RNA virus L2 | GaRV-L2 | YP_044806.1 | YP_044807.1 | |
| Helicobasidium mompa totivirus 1-17 | HmV1-17 | NP_898832.1 | NP_898833.1 | |
| Helminthosporium victoriae virus 190S | HvV190S | NP_619669.2 | NP_619670.2 | |
| Magnaporthe oryzae virus 1 | MoV1 | YP_122351.1 | YP_122352.1 | |
| Magnaporthe oryzae virus 2 | MoV2 | YP_001649205.1 | AB300379 | |
| Ustilaginoidea virens RNA virus 1 | UvRV1 | AGO04406.1 | AGO04407.1 | |
| Ustilaginoidea virens RNA virus 3 | UvRV3 | YP_009004155.1 | YP_009004156.1 | |
| Aspergillus foetidus slow virus 1 | AfSV1 | CCD33023.1 | CCD33024.1 | |
| Sclerotinia nivalis victorivirus 1 | SnV1 | YP_009259367.1 | YP_009259368.1 | |
| Nigrospora oryzae victorivirus 1 | NoV1 | YP_009254735.1 | YP_009254736.1 | |
| Sphaeropsis sapinea RNA virus 2 | SsRV2 | NP_047559.1 | NP_047560.1 | |
| Tolypocladium cylindrosporum virus 1 | TcV1 | YP_004089629.1 | YP_004089630.1 | |
| Phomopsis vexans RNA virus | PvRV | YP_009115491.1 | YP_009115492.1 | |
| Totivirus | Saccharomyces cerevisiae virus L-A | ScV-L-A | NP_620494.1 | AAA50321.1 |
| Tuber aestivum virus 1 | TaV1 | ADQ54105.1 | ADQ54106.1 | |
| Saccharomyces cerevisiae virus L-BC (La) | ScV-L-BC (La) | NP_042580.1 | NP_042581.1 | |
| Trichomonasvirus | Trichomonas vaginalis virus 1 | TVV1 | ABC86750.1 | ABC86751.1 |
| Trichomonas vaginalis virus 2 | TVV2 | AED99809.1 | AAF29445.1 | |
| Trichomonas vaginalis virus 3 | TVV3 | NP_659389.1 | NP_659390.1 | |
| Trichomonas vaginalis virus 4 | TVV4 | AED99797.1 | AED99798.1 | |
| Leishmania virus | Leishmania RNA virus 1-1 | LRV1-1 | NP_041190.1 | M92355 |
| Leishmania RNA virus 1-4 | LRV1-4 | NP_619652.1 | NP_619653.1 | |
| Leishmania RNA virus 2-1 | LRV2-1 | NP_043464.1 | U32108 | |
| Giardiavirus | Giardia lamblia virus | GLV | NP_619551.1 | NP_620070.1 |
| Unassigned | Eimeria brunetti RNA virus 1 | EbRV1 | NP_108650.1 | CAK02788.1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, J.; Cheng, C.Y.; Gao, B.D.; Zhou, Q.; Zhu, H.J. Mycoviruses in the Plant Pathogen Ustilaginoidea virens Are Not Correlated with the Genetic Backgrounds of Its Hosts. Int. J. Mol. Sci. 2017, 18, 963. https://doi.org/10.3390/ijms18050963
Zhong J, Cheng CY, Gao BD, Zhou Q, Zhu HJ. Mycoviruses in the Plant Pathogen Ustilaginoidea virens Are Not Correlated with the Genetic Backgrounds of Its Hosts. International Journal of Molecular Sciences. 2017; 18(5):963. https://doi.org/10.3390/ijms18050963
Chicago/Turabian StyleZhong, Jie, Chuan Yuan Cheng, Bi Da Gao, Qian Zhou, and Hong Jian Zhu. 2017. "Mycoviruses in the Plant Pathogen Ustilaginoidea virens Are Not Correlated with the Genetic Backgrounds of Its Hosts" International Journal of Molecular Sciences 18, no. 5: 963. https://doi.org/10.3390/ijms18050963





