The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs
Abstract
:1. Introduction
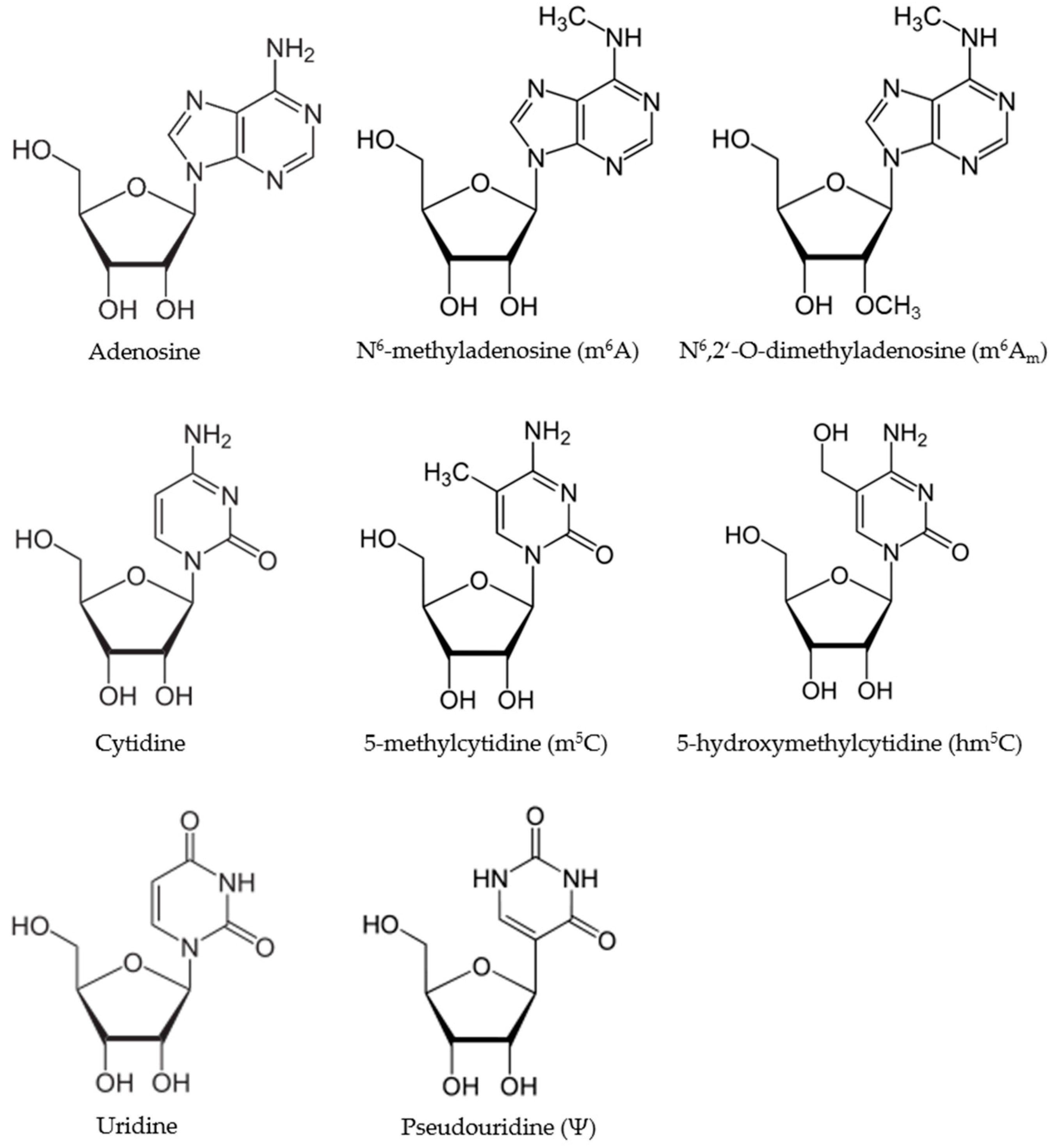
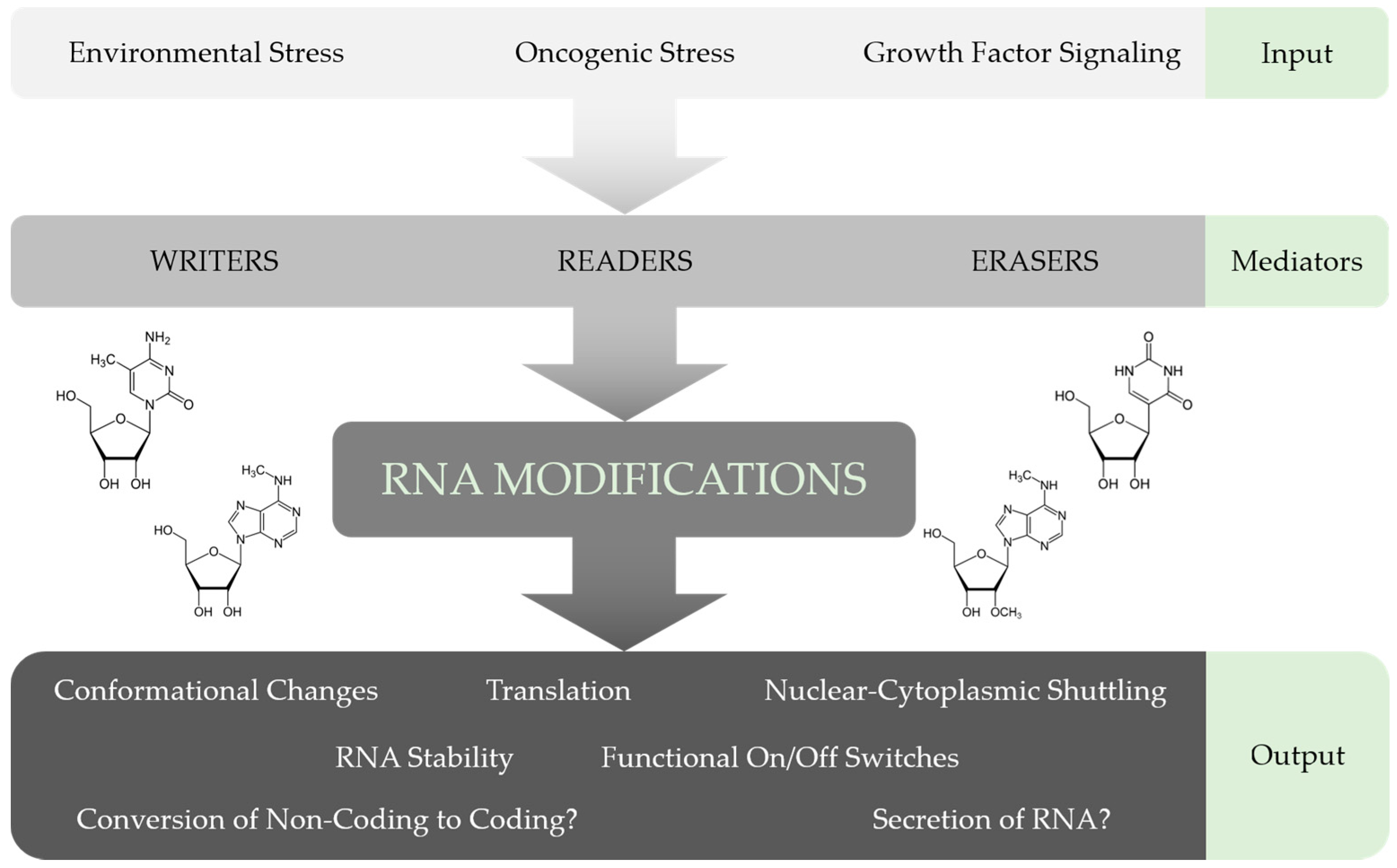
2. Discovery and Function of RNA Modifications
2.1. Pseudouridine
Ψ Detection Methods
2.2. N6-Methyladenosine
2.2.1. m6A Writers
2.2.2. m6A Readers
2.2.3. m6A Erasers
2.2.4. m6A Detection
2.3. 5-Methylcytosine
m5C Detection
3. RNA Modifications in Cancer-Related lncRNAs
3.1. MALAT1
3.2. HOTAIR
3.3. XIST
3.4. SRA1
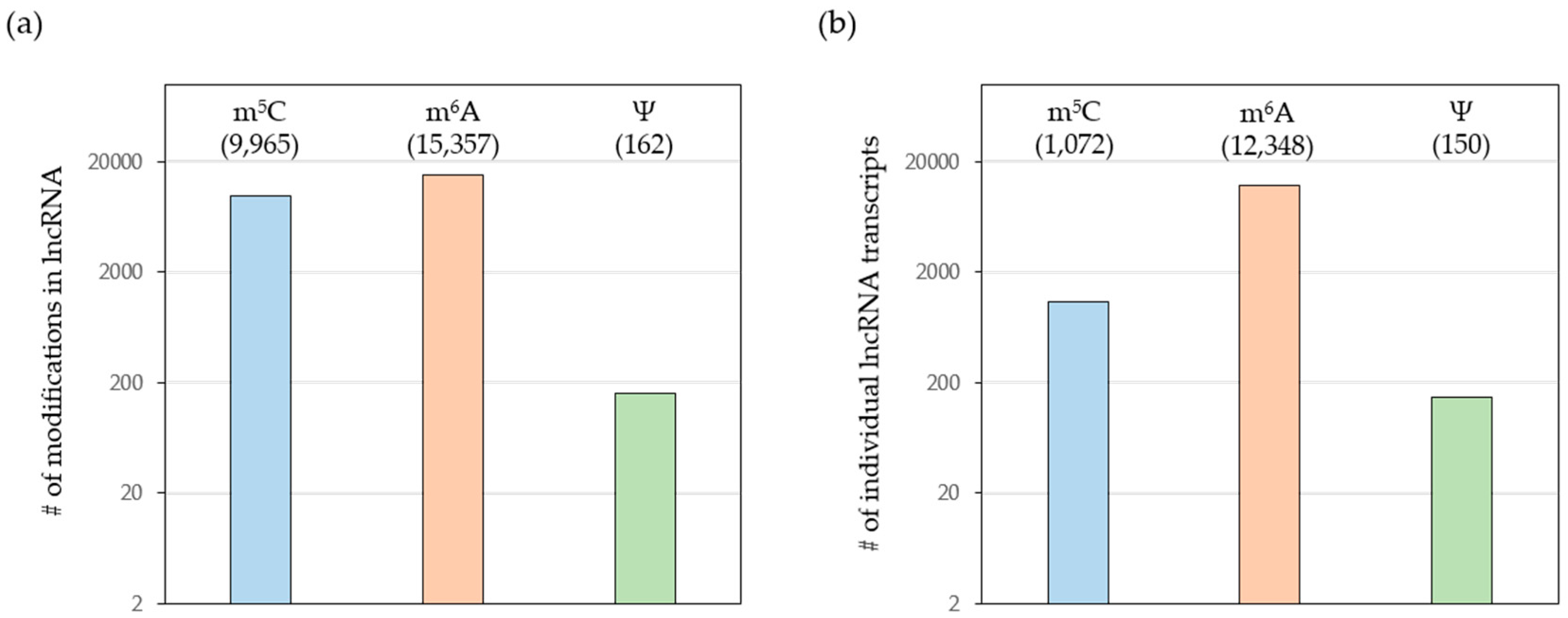
3.5. Additional lncRNAs with Posttranscriptional Chemical Modifications
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 4-SU | 4-thiouridine |
| ALKBH5 | α-ketoglutarate-dependent dioxygenase alkB homolog 5 |
| ALL | Acute lymphoblastic leukemia |
| ALYREF | Aly/REF export factor |
| AML | Acute myeloid leukemia |
| ANLL | Acute nonlymphoblastic leukemia |
| ANRIL | Antisense Non-coding RNA in the INK4 Locus |
| APTR | Alu-mediated CDKN1A/p21 transcriptional regulator |
| ARE | AU-rich element |
| Aza-IP | 5-azacytidine-mediated RNA immunoprecipitation |
| CeU-Seq | N3-CMC-enriched pseudouridine sequencing |
| CIMS | Cross-linking–induced mutation site |
| CITS | Cross-linking–induced truncation site |
| CMCT | N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate |
| CML | Chronic myeloid leukemia |
| CoREST | Co-repressor of RE1-silencing transcription factor |
| CRC | Colorectal cancer |
| DICER1-AS1 | DICER1 antisense RNA 1 |
| DKC1 | Dyskerin pseudouridine synthase 1 |
| DLEU2L | Deleted in lymphocytic leukemia 2-like |
| DNMT2 | DNA methyltransferase-2 |
| eIF3 | Eukaryotic initiation factor 3 |
| eIF4E | Eukaryotic initiation factor 4E |
| EMT | Epithelial-mesenchymal transition |
| FTO | Fat mass- and obesity-associated protein |
| GAS5 | Growth arrest specific 5 |
| GBM | Glioblastoma |
| GSC | Glioblastoma stem-like cells |
| H3K27 | Histone H3 lysine-27 |
| H3K4 | Histone H3 lysine-4 |
| hnRNP U | Heterogeneous nuclear ribonucleoprotein U |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 |
| HNRNPC | Heterogeneous nuclear ribonucleoprotein C |
| HOTAIR | HOX antisense intergenic RNA |
| HOXC/D | Homeobox C/D |
| HuR | Human antigen R |
| iCLIP | Individual-nucleotide-resolution crosslinking and immunoprecipitation |
| lncRNA | Long non-coding RNA |
| LRRC75C-AS1 | LRRC75A antisense RNA1 |
| LSD1 | Lysine specific demethylase 1A |
| m5C | 5-methylcytosine |
| m6A | N6-methyladenosine |
| m6Am | N6,2′-O-dimethyladenosine |
| MAGI2-AS3 | MAGI2 antisense RNA 3 |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 |
| mascRNA | MALAT1-associated small cytoplasmic RNA |
| MAT2A | Methionine adenosyltransferase 2A |
| MDS | Myelodysplastic syndrome |
| MeRIP-Seq | M6A specific methylated RNA immunoprecipitation-sequencing |
| METTL | Methyltransferase-like protein |
| miRNA | MicroRNA |
| MPN | Myeloproliferative neoplasm |
| mRNA | Messenger RNA |
| MS | Mass spectrometry |
| mtRNA | Mitochondrial RNA |
| ncRNA | Non-coding RNA |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 |
| NEAT2 | Nuclear-Enriched Abundant Transcript 2 |
| NGS | Next-generation sequencing |
| NSCLC | Non-small cell lung cancer |
| NSUN | NOP2/Sun RNA methyltransferase family member |
| nt | Nucleotide |
| PA-m6A-seq | Photo-crosslinking-assisted m6A sequencing |
| PcG | Polycomb group |
| piRNA | PIWI-interacting RNA |
| PRC2 | Polycomb-repressive complex 2 |
| PTM | Posttranslational modification |
| PUS | Pseudouridine synthase |
| PUS7L | Pseudouridylate synthase 7 like |
| PUSL1 | Pseudouridylate synthase-like 1 |
| PVT1 | Pvt1 oncogene |
| RAR | Retinoic acid receptor |
| RBP | RNA-binding protein |
| REST | RE1-silencing transcription factor |
| RBM15 | RNA-binding motif protein 15 |
| RMT | RNA methyltransferase |
| RN7SK | RNA 7SK small nuclear |
| RPPH1 | Ribonuclease P RNA component H1 |
| RPUSD | RNA pseudouridylate synthase domain containing |
| rRNA | Ribosomal RNA |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| SAM | S-Adenosyl methionine |
| SCARLET | Site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography |
| siRNA | Small interfering RNA |
| SNHG | Small nucleolar RNA host gene |
| snoRNA | Small nucleolar RNA |
| snRNA | Small nuclear RNA |
| SPEN | Spen family transcriptional repressor |
| SRA | Steroid receptor RNA activator |
| SRAP | Steroid receptor RNA activator protein |
| ST7-AS1 | ST7 antisense RNA 1 |
| TERC | Telomerase RNA component |
| tRNA | Transfer RNA |
| TRUB | TruB pseudouridine synthase family member |
| TUG1 | Taurine up-regulated 1 |
| UTR | Untranslated region |
| VIRMA | Vir like m6A methyltransferase associated |
| WTAP | Wilms’ tumor 1-associating protein |
| XIC | X inactivation center |
| XIST | X-inactive specific transcript |
| YTH | YT521-B homology |
| YTHDC | YTH domain-containing protein |
| YTHDF | YT521-B homology domain family |
| ZFAS1 | ZNFX1 antisense RNA 1 |
| Ψ | Pseudouridine |
References
- Prabakaran, S.; Lippens, G.; Steen, H.; Gunawardena, J. Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948, 175, 315–332. [Google Scholar] [PubMed]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Knowling, S.; Morris, K.V. Non-coding RNA and antisense RNA. Nature’s trash or treasure? Biochimie 2011, 93, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Klupp, F.; Brand, K.; Lasitschka, F.; Diederichs, S.; Kirchberg, J.; Rahbari, N.; Dutta, S.; Bork, U.; Fritzmann, J.; et al. Invasion front-specific expression and prognostic significance of microRNA in colorectal liver metastases. Cancer Sci 2011, 102, 1799–1807. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Roth, A.; Yu, M.; Morris, R.; Bersani, F.; Rivera, M.N.; Lu, J.; Shioda, T.; Vasudevan, S.; Ramaswamy, S.; et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013, 27, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Stiegelbauer, V.; Vychytilova-Faltejskova, P.; Ivan, C.; Ling, H.; Winter, E.; Zhang, X.; Goblirsch, M.; Wulf-Goldenberg, A.; Ohtsuka, M.; et al. Genome-Wide miRNA Analysis Identifies miR-188–3p as a Novel Prognostic Marker and Molecular Factor Involved in Colorectal Carcinogenesis. Clin. Cancer Res. 2017, 23, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, M.S.; Higgs, D.R.; Gingeras, T.R. Molecular biology: RNA discrimination. Nature 2012, 482, 310–311. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Carmichael, G.G. Long noncoding RNAs in mammalian cells: What, where, and why? Wiley Interdiscip. Rev. RNA 2010, 1, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Lipovich, L.; Johnson, R.; Lin, C.Y. MacroRNA underdogs in a microRNA world: Evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim. Biophys. Acta 2010, 1799, 597–615. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, S.; Diederichs, S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer 2016, 139, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Diederichs, S.; Bartsch, L.; Berkmann, J.C.; Frose, K.; Heitmann, J.; Hoppe, C.; Iggena, D.; Jazmati, D.; Karschnia, P.; Linsenmeier, M.; et al. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 2016, 8, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, M.; Gutschner, T. Long non-coding RNAs in cancer and development: Where do we go from here? Int. J. Mol. Sci. 2015, 16, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Pichler, M.; Chen, M.; Calin, G.A.; Ling, H. To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 2003. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucl. Acids Res. 2015, 43, D168–D173. [Google Scholar] [CrossRef] [PubMed]
- Cohn, W.E.; Volkin, E. Nucleoside-5’-Phosphates from Ribonucleic Acid. Nature 1951, 167, 483–484. [Google Scholar] [CrossRef]
- Wyatt, G.R. Occurrence of 5-methylcytosine in nucleic acids. Nature 1950, 166, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA Modifications: Playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways--2013 update. Nucl. Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucl. Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Decatur, W.A.; Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002, 27, 344–351. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Jaffrey, S.R.; Pan, T.; Rechavi, G.; Suzuki, T. RNA modifications: What have we learned and where are we headed? Nat. Rev. Genet. 2016, 17, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Patton, J.R. Cloning and characterization of a mammalian pseudouridine synthase. RNA 1999, 5, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Patton, J.R.; Davis, S.L.; Florence, B.; Ames, S.J.; Spanjaard, R.A. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell 2004, 15, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, S.; Yi, C. Pseudouridine: The fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 2016, 33, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Patton, J.R. Pseudouridine synthase 3 from mouse modifies the anticodon loop of tRNA. Biochemistry 2000, 39, 12723–12730. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Han, L.; Faqeih, E.; Ewida, N.; Alobeid, E.; Phizicky, E.M.; Alkuraya, F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016, 135, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Safra, M.; Nir, R.; Farouq, D.; Vainberg Slutskin, I.; Schwartz, S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017, 27, 393–406. [Google Scholar] [CrossRef] [PubMed]
- McCleverty, C.J.; Hornsby, M.; Spraggon, G.; Kreusch, A. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007, 373, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Antonicka, H.; Choquet, K.; Lin, Z.Y.; Gingras, A.C.; Kleinman, C.L.; Shoubridge, E.A. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 2017, 18, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Heiss, N.S.; Knight, S.W.; Vulliamy, T.J.; Klauck, S.M.; Wiemann, S.; Mason, P.J.; Poustka, A.; Dokal, I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998, 19, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Yang, J.; Watzinger, P.; Kotter, P.; Entian, K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucl. Acids Res. 2013, 41, 9062–9076. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Watt, F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006, 16, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Sloan, K.E.; Ranjan, N.; Warda, A.S.; Kretschmer, J.; Blessing, C.; Hubner, B.; Seikowski, J.; Dennerlein, S.; Rehling, P.; et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016, 35, 2104–2119. [Google Scholar] [CrossRef] [PubMed]
- Metodiev, M.D.; Spahr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.G.; Ruzzenente, B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014, 10, e1004110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haag, S.; Warda, A.S.; Kretschmer, J.; Gunnigmann, M.A.; Hobartner, C.; Bohnsack, M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 2015, 21, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.; Marquez, B.; Suarez, S.; Schimenti, J. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol. Reprod. 2007, 77, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Spenkuch, F.; Motorin, Y.; Helm, M. Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biol. 2014, 11, 1540–1554. [Google Scholar] [CrossRef] [PubMed]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [PubMed]
- Davis, D.R. Stabilization of RNA stacking by pseudouridine. Nucl. Acids Res. 1995, 23, 5020–5026. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yu, Y.T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011, 474, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Zaganelli, S.; Rebelo-Guiomar, P.; Maundrell, K.; Rozanska, A.; Pierredon, S.; Powell, C.A.; Jourdain, A.A.; Hulo, N.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.; et al. The Pseudouridine Synthase RPUSD4 Is an Essential Component of Mitochondrial RNA Granules. J. Biol. Chem. 2017, 292, 4519–4532. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.R.; Bykhovskaya, Y.; Mengesha, E.; Bertolotto, C.; Fischel-Ghodsian, N. Mitochondrial myopathy and sideroblastic anemia (MLASA): Missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 2005, 280, 19823–19828. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.P.; Liao, J.P.; Shen, J.; Yu, L.; Liu, B.L.; Liu, L.; Li, R.Y.; Ji, L.; Dorsey, S.G.; Jiang, Z.R.; et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012, 31, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Bykhovskaya, Y.; Casas, K.; Mengesha, E.; Inbal, A.; Fischel-Ghodsian, N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 2004, 74, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Yu, Y.T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013, 38, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Bakin, A.; Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry 1993, 32, 9754–9762. [Google Scholar] [CrossRef] [PubMed]
- Bakin, A.V.; Ofengand, J. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol. 1998, 77, 297–309. [Google Scholar] [PubMed]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; Leon-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, A.F.; Riordan, D.P.; Brown, P.O. Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 2014, 9, e110799. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, P.; Ma, S.; Song, J.; Bai, J.; Sun, F.; Yi, C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015, 11, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Parisien, M.; Dai, Q.; Zheng, G.; He, C.; Pan, T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 2013, 19, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, A.; Limbach, P.A. Mass spectrometry of the fifth nucleoside: A review of the identification of pseudouridine in nucleic acids. Anal. Chim. Acta 2008, 623, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Nobe, Y.; Izumikawa, K.; Higo, D.; Yamagishi, Y.; Takahashi, N.; Nakayama, H.; Isobe, T.; Taoka, M. A mass spectrometry-based method for direct determination of pseudouridine in RNA. Nucl. Acids Res. 2016, 44, e59. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.P.; Kelley, D.E. Existence of methylated messenger RNA in mouse L cells. Cell 1974, 1, 37–42. [Google Scholar] [CrossRef]
- Molinie, B.; Wang, J.; Lim, K.S.; Hillebrand, R.; Lu, Z.X.; van Wittenberghe, N.; Howard, B.D.; Daneshvar, K.; Mullen, A.C.; Dedon, P.; et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 2016, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.M.; Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977, 16, 1672–1676. [Google Scholar] [CrossRef] [PubMed]
- Csepany, T.; Lin, A.; Baldick, C.J., Jr.; Beemon, K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 1990, 265, 20117–20122. [Google Scholar] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Kierzek, E.; Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucl. Acids Res. 2003, 31, 4472–4480. [Google Scholar] [CrossRef] [PubMed]
- Roost, C.; Lynch, S.R.; Batista, P.J.; Qu, K.; Chang, H.Y.; Kool, E.T. Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J. Am. Chem. Soc. 2015, 137, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Laurent, B.; Hsu, C.H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Sledz, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. Elife 2016, 5, e18434. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [PubMed]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Lu, Z.; Wang, X.; Fu, Y.; Luo, G.-Z.; Liu, N.; Han, D.; Dominissini, D.; Dai, Q.; Pan, T.; et al. High-Resolution N6-Methyladenosine (m6A) Map Using Photo-Crosslinking-Assisted m6A Sequencing. Angew. Chem. 2015, 54, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Rieder, D.; Wille, A.; Khokhlova-Cubberley, D.; Riml, C.; Trixl, L.; Jia, X.Y.; Micura, R.; Lusser, A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucl. Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Brzezicha, B.; Schmidt, M.; Makalowska, I.; Jarmolowski, A.; Pienkowska, J.; Szweykowska-Kulinska, Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucl. Acids Res. 2006, 34, 6034–6043. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Abbasi-Moheb, L.; Mertel, S.; Gonsior, M.; Nouri-Vahid, L.; Kahrizi, K.; Cirak, S.; Wieczorek, D.; Motazacker, M.M.; Esmaeeli-Nieh, S.; Cremer, K.; et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Rafiq, M.A.; Noor, A.; Hussain, S.; Flores, J.V.; Rupp, V.; Vincent, A.K.; Malli, R.; Ali, G.; Khan, F.S.; et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 856–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, F.J.; Lee, J.H.; Lee, J.E.; Blanco, S.; Nickerson, E.; Gabriel, S.; Frye, M.; Al-Gazali, L.; Gleeson, J.G. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 2012, 49, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Gao, R.; Chen, Y.; Yang, Z.; Han, P.; Zhang, H.; Dou, Y.; Liu, W.; Wang, W.; Du, G.; et al. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget 2017, 8, 20751–20765. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Hirata, S.; Sato, S.; Koga, S.; Fujii, M.; Qi, G.; Ogawa, I.; Takata, T.; Shimamoto, F.; Tatsuka, M. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012, 31, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.; Jurkowski, T.P.; Kellner, S.; Schneider, D.; Jeltsch, A.; Helm, M. The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2016, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucl. Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef] [PubMed]
- Elhardt, W.; Shanmugam, R.; Jurkowski, T.P.; Jeltsch, A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie 2015, 112, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucl. Acids Res. 2009, 37, e12. [Google Scholar] [CrossRef] [PubMed]
- Shafik, A.; Schumann, U.; Evers, M.; Sibbritt, T.; Preiss, T. The emerging epitranscriptomics of long noncoding RNAs. Biochim. Biophys. Acta 2016, 1859, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat. Protoc. 2014, 9, 337–361. [Google Scholar] [CrossRef] [PubMed]
- Konig, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010, 17, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012, 2, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Baas, M.; Diederichs, S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011, 21, 1944–1954. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, Y.; Pang, J.; Weng, X.; Feng, X.; Guo, Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J. Cell Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Pang, E.J.; Yang, R.; Fu, X.B.; Liu, Y.F. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015, 36, 2403–2407. [Google Scholar] [CrossRef] [PubMed]
- Pa, M.; Naizaer, G.; Seyiti, A.; Kuerbang, G. Long Noncoding RNA MALAT1 Functions as a Sponge of MiR-200c in Ovarian Cancer. Oncol. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, X.; Yan, D.; Li, D.; Guan, F.; Dong, Y.; Wang, H.; Liu, X.; Yang, B. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell. Physiol. Biochem. 2017, 42, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Gast, M.; Schroen, B.; Voigt, A.; Haas, J.; Kuehl, U.; Lassner, D.; Skurk, C.; Escher, F.; Wang, X.; Kratzer, A.; et al. Long noncoding RNA MALAT1-derived mascRNA is involved in cardiovascular innate immunity. J. Mol. Cell Biol. 2016, 8, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 14013–14018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.I.; Parisien, M.; Dai, Q.; Liu, N.; Diatchenko, L.; Sachleben, J.R.; Pan, T. N6-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding. J. Mol. Biol. 2016, 428, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Song, X.Q.; Cai, J.P.; Zhang, S. HOTAIR: A cancer-related long non-coding RNA. Neoplasma 2014, 61, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
- Richtig, G.; Ehall, B.; Richtig, E.; Aigelsreiter, A.; Gutschner, T.; Pichler, M. Function and Clinical Implications of Long Non-Coding RNAs in Melanoma. Int. J. Mol. Sci. 2017, 18, 715. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, L.; Zheng, S. Role of the long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol. Lett. 2017, 14, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Kong, D.; Chen, Q.; Ping, Y.; Pang, D. Oncogenic long noncoding RNA landscape in breast cancer. Mol. Cancer 2017, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Y.; Liu, X.; Luo, P.; Jing, W.; Zhu, M.; Tu, J. Identification of Circulating Long Noncoding RNA HOTAIR as a Novel Biomarker for Diagnosis and Monitoring of Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Souliere, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Worle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011, 12, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, S.T.; Heard, E. Novel players in X inactivation: Insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 2017, 24, 197–204. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Minajigi, A.; Froberg, J.; Wei, C.; Sunwoo, H.; Kesner, B.; Colognori, D.; Lessing, D.; Payer, B.; Boukhali, M.; Haas, W.; et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015, 349. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, E.; Kirby, J.E.; Brown, D.E.; Mercier, F.E.; Sadreyev, R.I.; Scadden, D.T.; Lee, J.T. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 2013, 152, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.M.; Orazi, A. Diagnostic criteria to distinguish hypocellular acute myeloid leukemia from hypocellular myelodysplastic syndromes and aplastic anemia: Recommendations for a standardized approach. Haematologica 2009, 94, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Dewald, G.W.; Brecher, M.; Travis, L.B.; Stupca, P.J. Twenty-six patients with hematologic disorders and X chromosome abnormalities. Frequent idic(X)(q13) chromosomes and Xq13 anomalies associated with pathologic ringed sideroblasts. Cancer Genet. Cytogenet. 1989, 42, 173–185. [Google Scholar] [CrossRef]
- Dierlamm, J.; Michaux, L.; Criel, A.; Wlodarska, I.; Zeller, W.; Louwagie, A.; Michaux, J.L.; Mecucci, C.; van den Berghe, H. Isodicentric (X)(q13) in haematological malignancies: Presentation of five new cases, application of fluorescence in situ hybridization (FISH) and review of the literature. Br. J. Haematol. 1995, 91, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, K.; Haferlach, C.; Fonatsch, C.; Hagemeijer, A.; Andersen, M.K.; Slovak, M.L.; Johansson, B.; Foundation, M.D.S. The idic(X)(q13) in myeloid malignancies: Breakpoint clustering in segmental duplications and association with TET2 mutations. Hum. Mol. Genet. 2010, 19, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Rack, K.A.; Chelly, J.; Gibbons, R.J.; Rider, S.; Benjamin, D.; Lafreniere, R.G.; Oscier, D.; Hendriks, R.W.; Craig, I.W.; Willard, H.F.; et al. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet. 1994, 3, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, K.; Mahlamaki, E.; Riikonen, P.; Meltoranta, R.L.; Rahiala, J.; Perkkio, M. Acquired X-chromosome aneuploidy in children with acute lymphoblastic leukemia. Med. Pediatr. Oncol. 1999, 32, 360–365. [Google Scholar] [CrossRef]
- Yamamoto, K.; Nagata, K.; Kida, A.; Hamaguchi, H. Acquired gain of an X chromosome as the sole abnormality in the blast crisis of chronic neutrophilic leukemia. Cancer Genet. Cytogenet. 2002, 134, 84–87. [Google Scholar] [CrossRef]
- Lassmann, S.; Weis, R.; Makowiec, F.; Roth, J.; Danciu, M.; Hopt, U.; Werner, M. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J. Mol. Med. 2007, 85, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Chen, L.Z.; Lu, Y.X.; Zhang, D.S.; Zeng, Z.L.; Pan, Z.Z.; Huang, P.; Wang, F.H.; Li, Y.H.; Ju, H.Q.; et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017, 8, e3011. [Google Scholar] [CrossRef] [PubMed]
- Leygue, E. Steroid receptor RNA activator (SRA1): Unusual bifaceted gene products with suspected relevance to breast cancer. Nucl. Recept. Signal. 2007, 5, e006. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.B.; Chua, S.S.; Barron, N.; Soder, B.M.; DeMayo, F.; O Malley, B.W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell. Biol. 2003, 23, 7163–7176. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.B.; McKenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17–27. [Google Scholar] [CrossRef]
- Hube, F.; Guo, J.; Chooniedass-Kothari, S.; Cooper, C.; Hamedani, M.K.; Dibrov, A.A.; Blanchard, A.A.; Wang, X.; Deng, G.; Myal, Y.; et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006, 25, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.B.; Razani, B.; Goldberg, A.D.; O’Malley, B.W. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc. Natl. Acade. Sci. USA 2002, 99, 16081–16086. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, H.T.; Zhu, N.; Shi, Y.N.; Liu, Z.; Ao, B.X.; Liao, D.F.; Zheng, X.L.; Qin, L. Steroid receptor RNA activator: Biologic function and role in disease. Clin. Chim. Acta 2016, 459, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Patton, J.R.; Ghosh, S.K.; Fischel-Ghodsian, N.; Shen, L.; Spanjaard, R.A. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007, 21, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.K.; Theimer, C.A.; Mitchell, J.R.; Collins, K.; Feigon, J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucl. Acids Res. 2010, 38, 6746–6756. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Glasner, H.; Riml, C.; Micura, R.; Breuker, K. Label-free, direct localization and relative quantitation of the RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass spectrometry. Nucl. Acids Res. 2017, 45, 8014–8025. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.E.; Quinn, R.; Sayre, J.L.; Fabris, D. Profiling ribonucleotide modifications at full-transcriptome level: A step toward MS-based epitranscriptomics. RNA 2015, 21, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; Mackowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Calviello, L.; Mukherjee, N.; Wyler, E.; Zauber, H.; Hirsekorn, A.; Selbach, M.; Landthaler, M.; Obermayer, B.; Ohler, U. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 2016, 13, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.; Jackson, S.E.; Wills, M.R.; Weissman, J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Rahim, A.; Guigo, R.; Vardy, L.A.; Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016, 22, 867–882. [Google Scholar] [CrossRef] [PubMed]
| RNA Modification | Writer | Reader | Eraser |
|---|---|---|---|
| Ψ | PUS1 [39,40] PUSL1 [41] PUS3 [42,43] PUS7 [44] PUS7L [41] PUS10 [45] RPUSD1 [41] RPUSD2 [41] RPUSD3 [46] RPUSD4 [46] TRUB1 [44] TRUB2 [46] DKC1 [47] | ||
| m6A | METTL3 [48] METTL14 [49,50,51] WTAP [49,50,51,52] KIAA1429 [51,53] RBM15 [53,54] RBM15B [54] METTL16 [55] | YTHDF1 [56] YTHDF2 [57] YTHDF3 [57] YTHDC1 [58] YTHDC2 [54] eIF3 [59] HNRNPA2B1 [60] HNRNPC [61] | ALKBH5 [62] FTO [63] |
| m5C | NSUN1 [64] NSUN2 [65] NSUN3 [66] NSUN4 [67] NSUN5 [68] NSUN6 [69] NSUN7 [70] DNMT2 [71] | ALYREF [72] |
| lncRNA | Modification | No. of Modified Residues | Reference |
|---|---|---|---|
| ANRIL | m6A | 1 | [97] |
| m5C | 2 | [118] | |
| DICER1-AS1 | m6A | 2 | [97] |
| Ψ | 1 | [87] | |
| GAS5 | m5C | 2 | [118] |
| HOTAIR | m6A | 1 | [97] |
| m5C | 1 | [163] | |
| Kcnq1ot1 | Ψ | 1 | [87] |
| LRRC75A-AS1 | Ψ | 1 | [77] |
| MALAT1 | m6A | 3 | [57] |
| 3 | [97] | ||
| m5C | 7 | [118] | |
| Ψ | 3 | [77] | |
| 3 | [87] | ||
| NEAT1 | m6A | 1 | [57] |
| m5C | 7 | [118] | |
| PVT1 | m6A | 2 | [57] |
| 1 | [97] | ||
| m5C | 1 | [118] | |
| 1 | [120] | ||
| Ψ | 1 | [77] | |
| RPPH1 | m5C | 4 | [118] |
| 1 | [127] | ||
| 1 | [120] | ||
| SNHG1 | Ψ | 1 | [77] |
| SNHG7 | Ψ | 1 | [87] |
| SNHG12 | m5C | 2 | [118] |
| SRA1 | m6A | 1 | [57] |
| 4 | [97] | ||
| Ψ | 1 | [40,189] | |
| ST7-AS1 | Ψ | 1 | [87] |
| TERC | m5C | 3 | [118] |
| Ψ | 2 | [85] | |
| 6 | [190] | ||
| TUG1 | m6A | 1 | [88] |
| 11 | [97] | ||
| XIST | m6A | 1 | [57] |
| 14 | [97] | ||
| m5C | 5 | [163] | |
| Ψ | 1 | [87] | |
| ZFAS1 | Ψ | 1 | [85] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacob, R.; Zander, S.; Gutschner, T. The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs. Int. J. Mol. Sci. 2017, 18, 2387. https://doi.org/10.3390/ijms18112387
Jacob R, Zander S, Gutschner T. The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs. International Journal of Molecular Sciences. 2017; 18(11):2387. https://doi.org/10.3390/ijms18112387
Chicago/Turabian StyleJacob, Roland, Sindy Zander, and Tony Gutschner. 2017. "The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs" International Journal of Molecular Sciences 18, no. 11: 2387. https://doi.org/10.3390/ijms18112387




