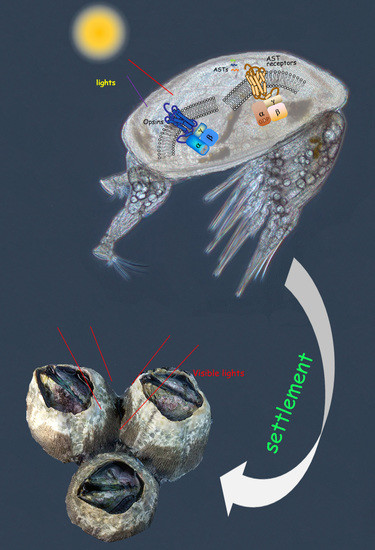
Comparative Transcriptomic Analysis Reveals Candidate Genes and Pathways Involved in Larval Settlement of the Barnacle Megabalanus volcano
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Sequencing, De Novo Assembly, and Unigene Annotation
2.2. Classification of Unigenes
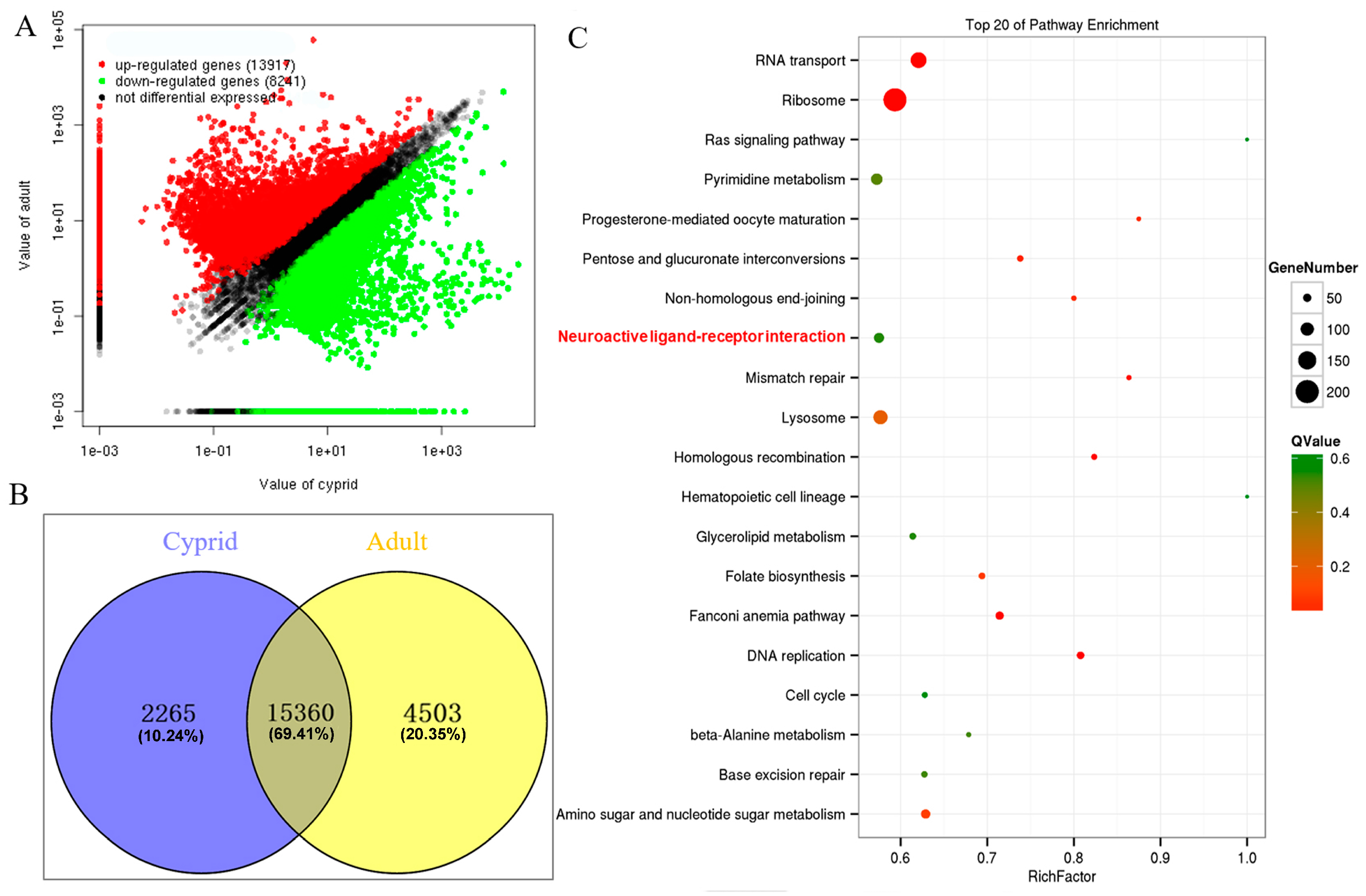
2.3. Identification and Enrichment Analysis of Differentially Expressed Genes
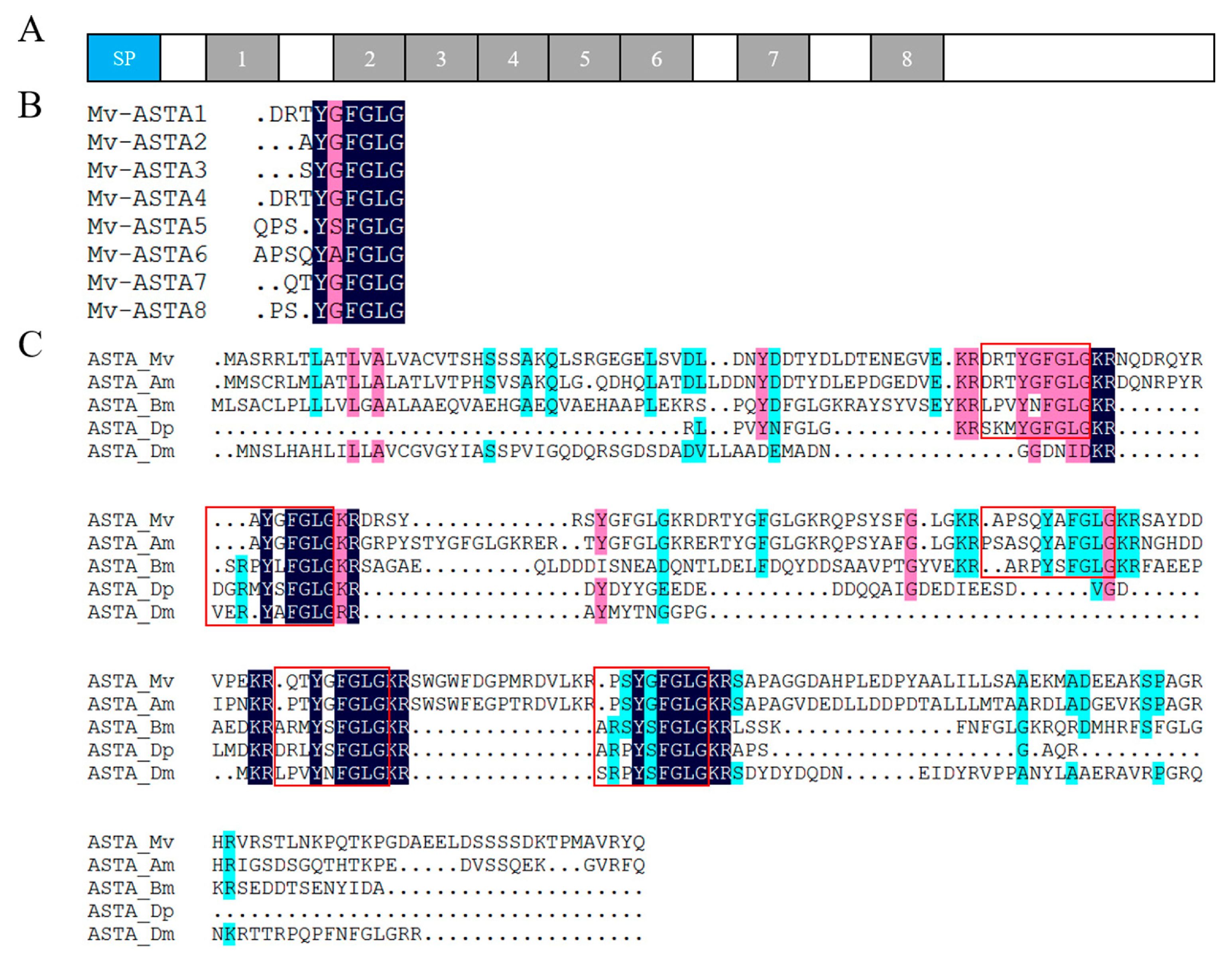
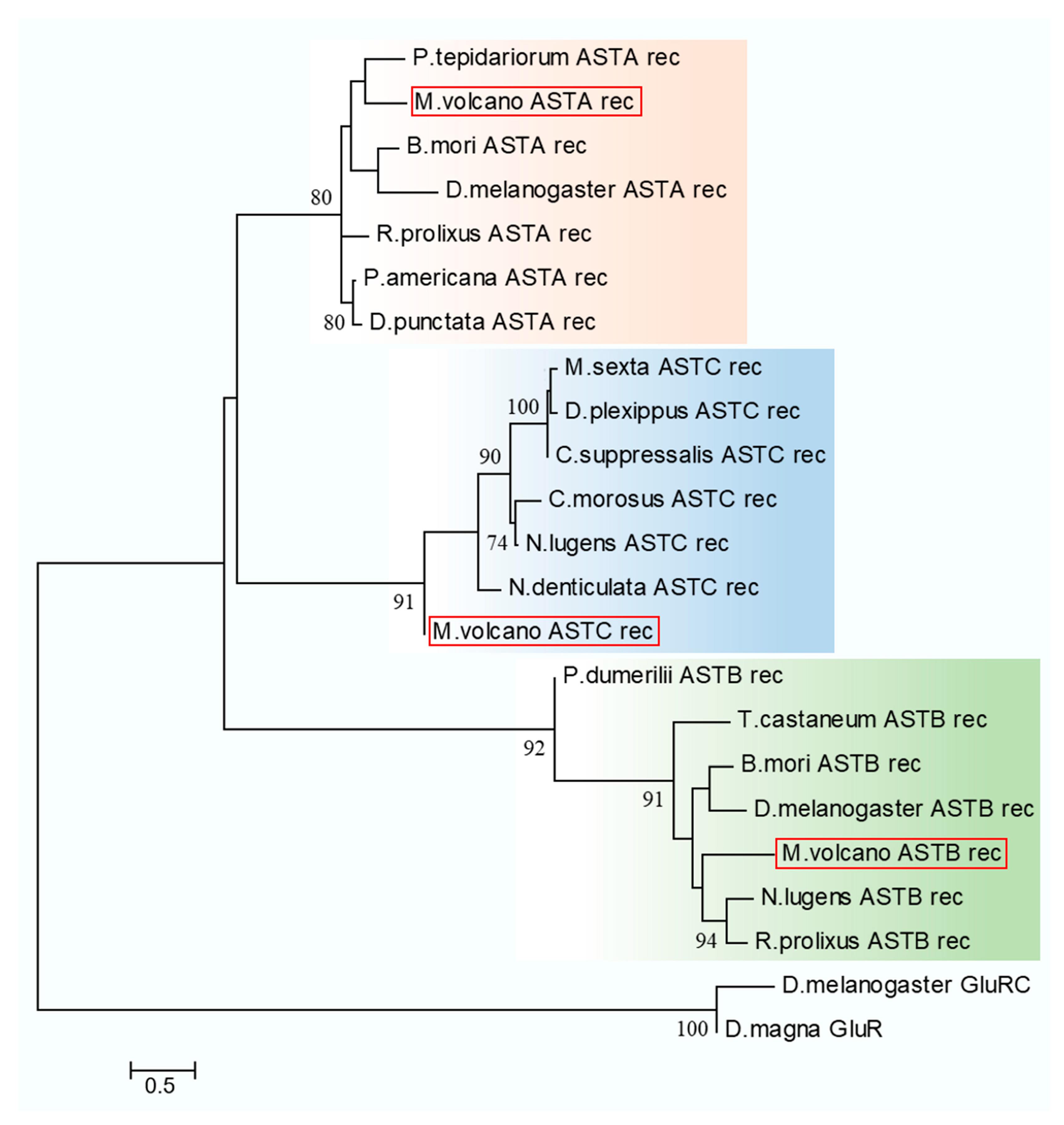
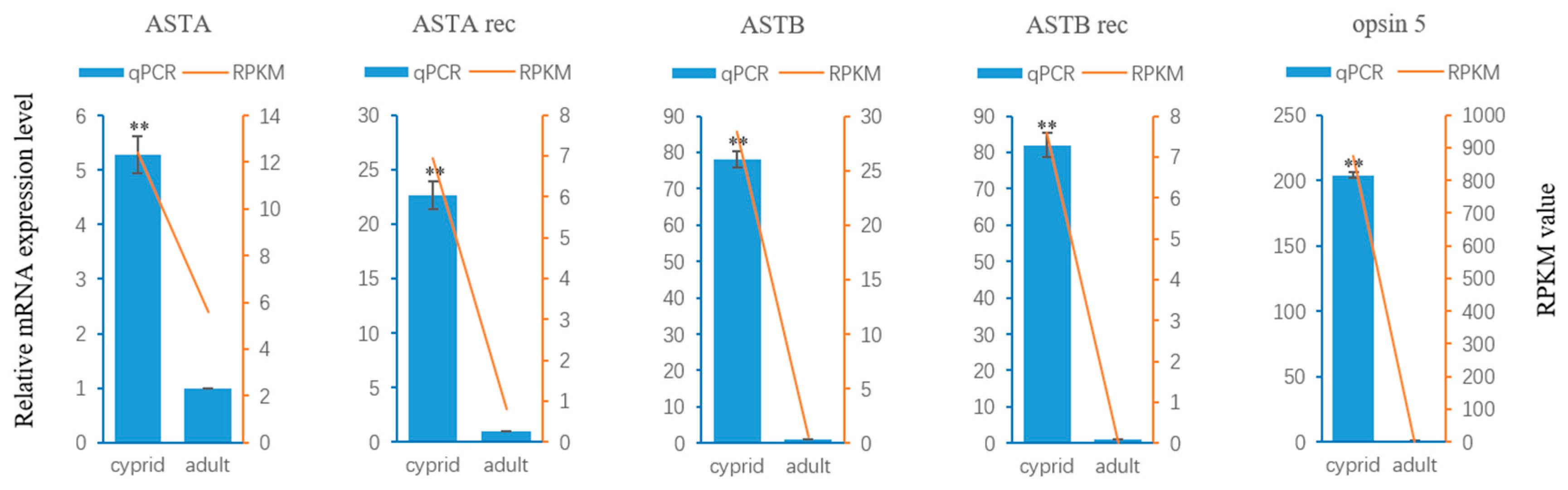
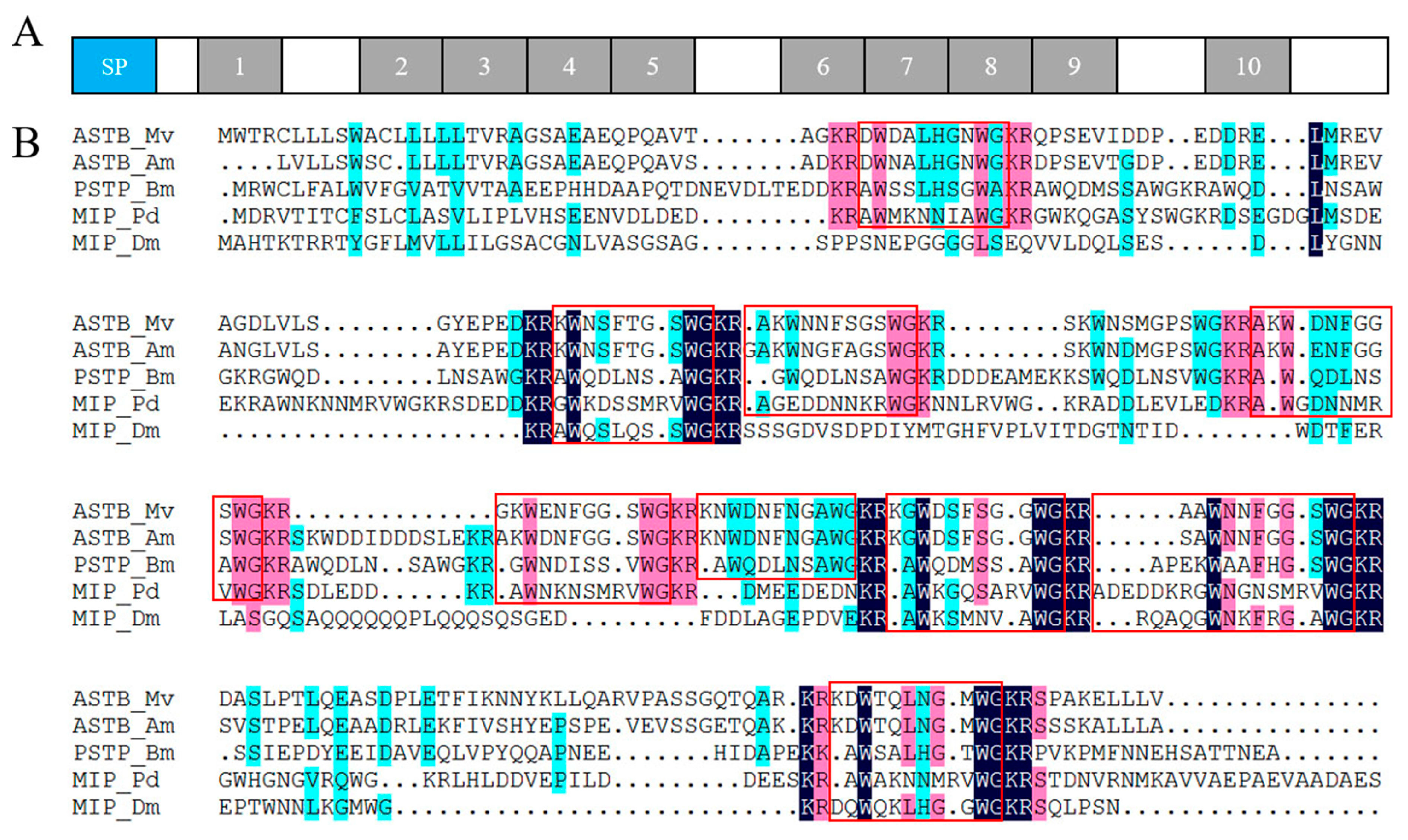
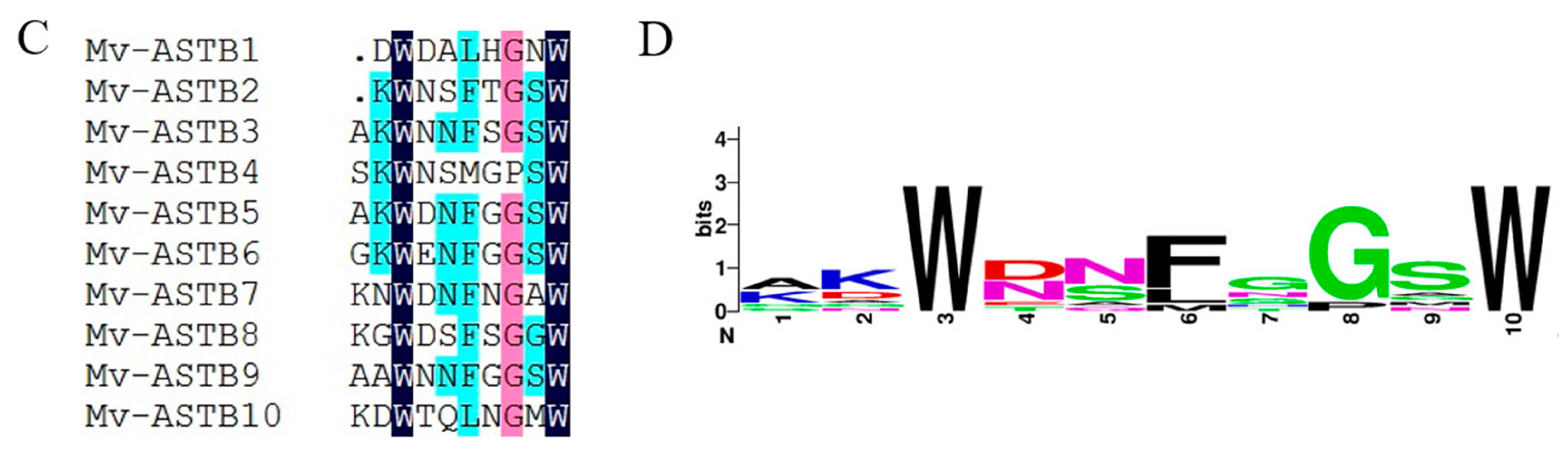
2.4. Neuropeptide Allatostatin Family and Their Putative Receptors
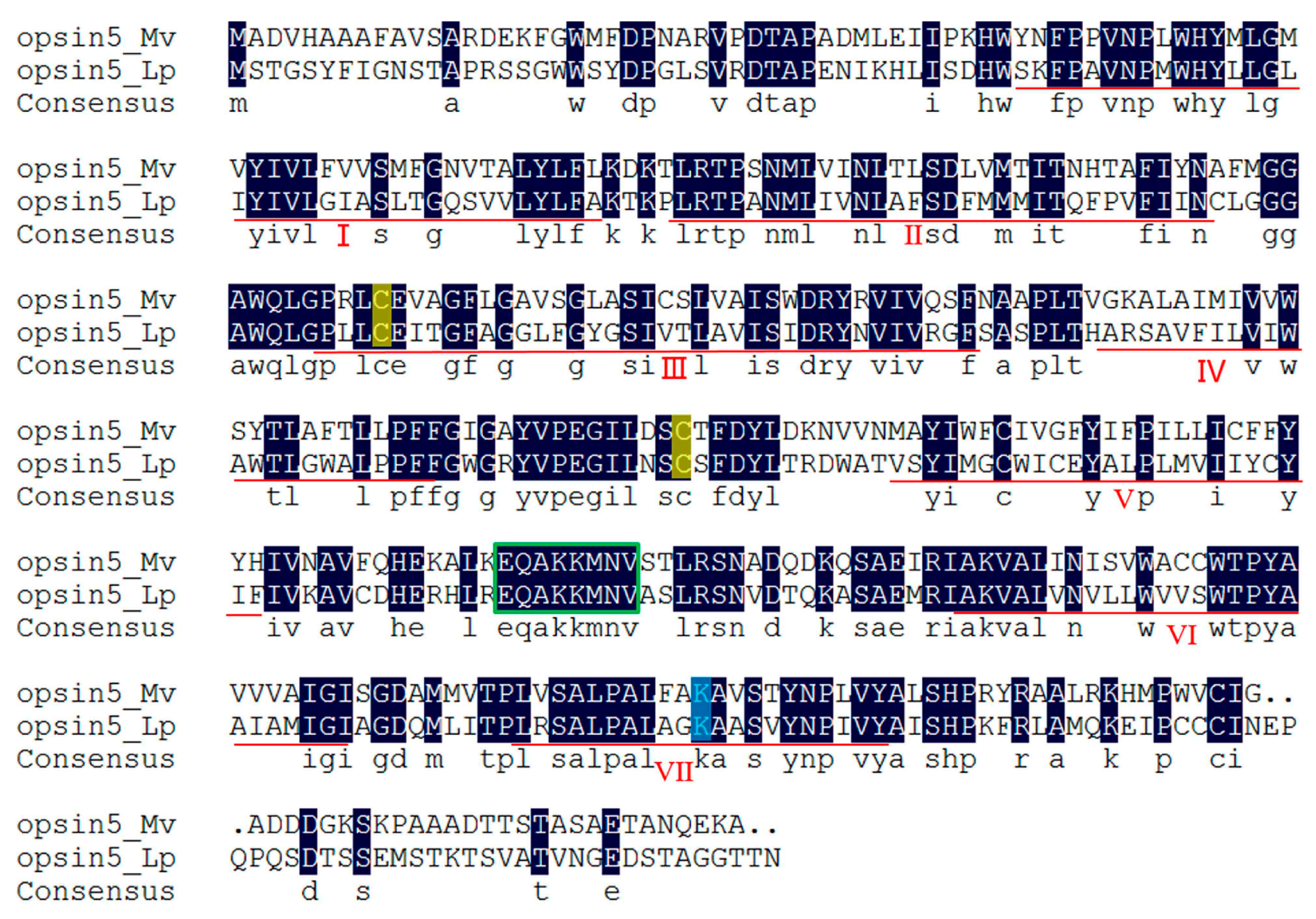
2.5. Light-Sensitive Proteins
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Larval Culture
4.3. RNA Extraction, RNA-Seq Library Construction and Sequencing
4.4. De Novo Assembly and Functional Annotation
4.5. Differential Gene Expression Analysis
4.6. Validation of RNA-Seq Results by Quantitative Real-Time PCR
4.7. Sequence Alignment and Phylogenetic Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jonker, J.L.; Morrison, L.; Lynch, E.P.; Grunwald, I.; von Byern, J.; Power, A.M. The chemistry of stalked barnacle adhesive (Lepas anatifera). Interface Focus 2015, 5, 20140062. [Google Scholar] [CrossRef] [PubMed]
- Fitridge, I.; Dempster, T.; Guenther, J.; de Nys, R. The impact and control of biofouling in marine aquaculture: A review. Biofouling 2012, 28, 649–669. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.P.; Bendick, J.A.; Holm, E.R.; Hertel, W.M. Economic impact of biofouling on a naval surface ship. Biofouling 2011, 27, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Clare, A.S.; Thomas, R.F.; Rittschof, D. Evidence for the involvement of cyclic-amp in the pheromonal modulation of barnacle settlement. J. Exp. Biol. 1995, 198, 655–664. [Google Scholar] [PubMed]
- Dreanno, C.; Matsumura, K.; Dohmae, N.; Takio, K.; Hirota, H.; Kirby, R.R.; Clare, A.S. An α(2)-macroglobulin-like protein is the cue to gregarious settlement of the barnacle Balanus amphitrite. Proc. Natl. Acad. Sci. USA 2006, 103, 14396–14401. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-F.; Matsumura, K.; Wang, H.; Arellano, S.M.; Yan, X.; Alam, I.; Archer, J.A.C.; Bajic, V.B.; Qian, P.-Y. Toward an understanding of the molecular mechanisms of barnacle larval settlement: A comparative transcriptomic approach. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.-C.; Chen, Z.-F.; Sun, J.; Matsumura, K.; Wu, R.S.S.; Qian, P.-Y. Transcriptomic analysis of neuropeptides and peptide hormones in the barnacle Balanus amphitrite: Evidence of roles in larval settlement. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.-S.; Xu, Y.; Matsumura, K.; Zhang, Y.; Zhang, G.; Qi, S.-H.; Qian, P.-Y. Evidence for the involvement of p38 MAPK activation in barnacle larval settlement. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; He, L.-S.; Wong, Y.H.; Qian, P.-Y. Mkk3 was involved in larval settlement of the barnacle amphibalanus amphitrite through activating the kinase activity of p38 mapk. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Wang, X.; Xiong, M.; Lei, C.; Zhu, F. The developmental transcriptome of the synanthropic fly Chrysomya megacephala and insights into olfactory proteins. BMC Genom. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.J.; Burton, K.J.; Zantello, M.R.; Smith, V.G.; Lowery, D.L.; Kubiak, T.M. Type a allatostatins from Drosophila melanogaster and Diplotera puncata activate two Drosophila allatostatin receptors, dar-1 and dar-2, expressed in CHO cells. Biochem. Biophys. Res. Commun. 2001, 286, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Secher, T.; Lenz, C.; Cazzamali, G.; Sorensen, G.; Williamson, M.; Hansen, G.N.; Svane, P.; Grimmelikhuijzen, C.J. Molecular cloning of a functional allatostatin gut/brain receptor and an allatostatin preprohormone from the silkworm Bombyx mori. J. Biol. Chem. 2001, 276, 47052–47060. [Google Scholar] [CrossRef] [PubMed]
- Zandawala, M.; Orchard, I. Identification and functional characterization of fglamide-related allatostatin receptor in Rhodnius prolixus. Insect Biochem. Mol. Biol. 2015, 57, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Bartalska, K.; Audsley, N.; Yamanaka, N.; Yapici, N.; Lee, J.Y.; Kim, Y.C.; Markovic, M.; Isaac, E.; Tanaka, Y.; et al. MIPs are ancestral ligands for the sex peptide receptor. Proc. Natl. Acad. Sci. USA 2010, 107, 6520–6525. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, N.; Hua, Y.J.; Roller, L.; Spalovska-Valachova, I.; Mizoguchi, A.; Kataoka, H.; Tanaka, Y. Bombyx prothoracicostatic peptides activate the sex peptide receptor to regulate ecdysteroid biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 2060–2065. [Google Scholar] [CrossRef] [PubMed]
- Conzelmann, M.; Williams, E.A.; Tunaru, S.; Randel, N.; Shahidi, R.; Asadulina, A.; Berger, J.; Offermanns, S.; Jekely, G. Conserved MIP receptor-ligand pair regulates Platynereis larval settlement. Proc. Natl. Acad. Sci. USA 2013, 110, 8224–8229. [Google Scholar] [CrossRef] [PubMed]
- Paluzzi, J.P.; Haddad, A.S.; Sedra, L.; Orchard, I.; Lange, A.B. Functional characterization and expression analysis of the myoinhibiting peptide receptor in the chagas disease vector, Rhodnius prolixus. Mol. Cell. Endocrinol. 2015, 399, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Katti, C.; Kempler, K.; Porter, M.L.; Legg, A.; Gonzalez, R.; Garcia-Rivera, E.; Dugger, D.; Battelle, B.A. Opsin co-expression in Limulus photoreceptors: Differential regulation by light and a circadian clock. J. Exp. Biol. 2010, 213, 2589–2601. [Google Scholar] [CrossRef] [PubMed]
- Kashiyama, K.; Seki, T.; Numata, H.; Goto, S.G. Molecular characterization of visual pigments in Branchiopoda and the evolution of opsins in Arthropoda. Mol. Biol. Evol. 2009, 26, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Futahashi, R.; Kawahara-Miki, R.; Kinoshita, M.; Yoshitake, K.; Yajima, S.; Arikawa, K.; Fukatsu, T. Extraordinary diversity of visual opsin genes in dragonflies. Proc. Natl. Acad. Sci. USA 2015, 112, E1247–E1256. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, C.R.; Fujimoto, M.S.; Lord, N.P.; Shin, S.; McKenna, D.D.; Suvorov, A.; Martin, G.J.; Bybee, S.M. Overcoming the loss of blue sensitivity through opsin duplication in the largest animal group, beetles. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Yan, T.; Li, Z.F.; Li, J.; Cheng, Z.Q. Fouling acorn barnacles in china-a review. Chin. J. Oceanol. Limn. 2013, 31, 699–711. [Google Scholar] [CrossRef]
- Hadfield, M.G.; Paul, V.J. Natural chemical cues for settlement and metamorphosis of marine invertebrate larvae. Mar. Chem. Ecol. 2001, 431–461. [Google Scholar] [CrossRef]
- Prendergast, G.S.; Zurn, C.M.; Bers, A.V.; Head, R.M.; Hansson, L.J.; Thomason, J.C. The relative magnitude of the effects of biological and physical settlement cues for cypris larvae of the acorn barnacle, Semibalanus balanoides L. Biofouling 2009, 25, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.J.H.; Sandeman, D.C. Morphology of the nervous system of the barnacle cypris larva (Balanus amphitrite darwin) revealed by light and electron microscopy. Biol. Bull. 1999, 197, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Gu, G.X.; Teng, Z.W.; Wu, S.F.; Huang, J.; Song, Q.S.; Ye, G.Y.; Fang, Q. Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Stay, B.; Tobe, S.S. The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annu. Rev. Entomol. 2007, 52, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.W.; Kellner, R.; Hoffmann, K.H. Identification of two allatostatins from the cricket, Gryllus bimaculatus de Geer (Ensifera, Gryllidae): Additional members of a family of neuropeptides inhibiting juvenile hormone biosynthesis. Regul. Pept. 1995, 57, 227–236. [Google Scholar] [CrossRef]
- Smith, P.A.; Clare, A.S.; Rees, H.H.; Prescott, M.C.; Wainwright, G.; Thorndyke, M.C. Identification of methyl farnesoate in the cypris larva of the barnacle, Balanus amphitrite, and its role as a juvenile hormone. Insect Biochem. Mol. Biol. 2000, 30, 885–890. [Google Scholar] [CrossRef]
- Yamamoto, H.; Okino, T.; Yoshimura, E.; Tachibana, A.; Shimizu, K.; Fusetani, N. Methyl farnesoate induces larval metamorphosis of the barnacle, Balanus amphitrite via protein kinase c activation. J. Exp. Zool. 1997, 278, 349–355. [Google Scholar] [CrossRef]
- Holland, D.; Walker, G. The biochemical composition of the cypris larva of the barnacle Balanus balanoides L. Ices J. Mar. Sci. 1975, 36, 162–165. [Google Scholar] [CrossRef]
- Lucas, M.; Walker, G.; Holland, D.; Crisp, D. An energy budget for the free-swimming and metamorphosing larvae of Balanus balanoides (crustacea: Cirripedia). Mar. Biol. 1979, 55, 221–229. [Google Scholar] [CrossRef]
- Walker, G.; Yule, A.; Nott, J. Structure and function in Balanomorph larvae. Crustac. Issues 1987, 5, 307–328. [Google Scholar]
- Hentze, J.L.; Carlsson, M.A.; Kondo, S.; Nassel, D.R.; Rewitz, K.F. The neuropeptide allatostatin a regulates metabolism and feeding decisions in Drosophila. Sci. Rep. 2015, 5, 11680. [Google Scholar] [CrossRef] [PubMed]
- Anil, A.C.; Khandeparker, L.; Desai, D.V.; Baragi, L.V.; Gaonkar, C.A. Larval development, sensory mechanisms and physiological adaptations in acorn barnacles with special reference to Balanus amphitrite. J. Exp. Mar. Biol. Ecol. 2010, 392, 89–98. [Google Scholar] [CrossRef]
- Matsumura, K.; Qian, P.Y. Larval vision contributes to gregarious settlement in barnacles: Adult red fluorescence as a possible visual signal. J. Exp. Biol. 2014, 217, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Eakin, R.M. Evolutionary significance of photoreceptors—Retrospect. Am. Zool. 1979, 19, 647–653. [Google Scholar] [CrossRef]
- Brierley, A.S. Diel vertical migration. Curr. Biol. 2014, 24, R1074–R1076. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, H.; Chen, I.S.; Kubo, Y.; Furutani, Y. A ciliary opsin in the brain of a marine annelid zooplankton is UV-sensitive and the sensitivity is tuned by a single amino acid residue. J. Biol. Chem. 2017, 292, 12971–12980. [Google Scholar] [CrossRef] [PubMed]
- Tapia, F.J.; DiBacco, C.; Jarrett, J.; Pineda, J. Vertical distribution of barnacle larvae at a fixed nearshore station in southern California: Stage-specific and diel patterns. Estuar. Coast. Shelf Sci. 2010, 86, 265–270. [Google Scholar] [CrossRef]
- Bonicelli, J.; Tyburczy, J.; Tapia, F.J.; Finke, G.R.; Parrague, M.; Dudas, S.; Menge, B.A.; Navarrete, S.A. Diet vertical migration and cross-shore distribution of barnacle and bivalve larvae in the central chile inner-shelf. J. Exp. Mar. Biol. Ecol. 2016, 485, 35–46. [Google Scholar] [CrossRef]
- Williamson, C.E.; Fischer, J.M.; Bollens, S.M.; Overholt, E.P.; Breckenridge, J.K. Toward a more comprehensive theory of zooplankton diel vertical migration: Integrating ultraviolet radiation and water transparency into the biotic paradigm. Limnol. Oceanogr. 2011, 56, 1603–1623. [Google Scholar] [CrossRef]
- Wacker, D.; Stevens, R.C.; Roth, B.L. How ligands illuminate gpcr molecular pharmacology. Cell 2017, 170, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Audsley, N.; Down, R.E. G protein coupled receptors as targets for next generation pesticides. Insect. Biochem. Mol. Biol. 2015, 67, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Dahlstrom, M.; Lindgren, F.; Berntsson, K.; Sjogren, M.; Martensson, L.G.; Jonsson, P.R.; Elwing, H. Evidence for different pharmacological targets for imidazoline compounds inhibiting settlement of the barnacle Balanus improvisus. J. Exp. Zool. Part A Comp. Exp. Biol. 2005, 303, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Lind, U.; Rosenblad, M.A.; Frank, L.H.; Falkbring, S.; Brive, L.; Laurila, J.M.; Pohjanoksa, K.; Vuorenpaa, A.; Kukkonen, J.P.; Gunnarsson, L.; et al. Octopamine receptors from the barnacle Balanus improvisus are activated by the α(2)-adrenoceptor agonist medetomidine. Mol. Pharmacol. 2010, 78, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Thiyagarajan, V.; Harder, T.; Qian, P.Y. Relationship between cyprid energy reserves and metamorphosis in the barnacle Balanus amphitrite darwin (Cirripedia; Thoracica). J. Exp. Mar. Biol. Ecol. 2002, 280, 79–93. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. Busco: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. Estscan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Heidelberg, Germany, 6–10 August 1999; pp. 138–148. [Google Scholar]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Cao, X.; Xu, X.; Huang, S.; Liu, C.; Tomljanovic, T. Developmental transcriptome analysis and identification of genes involved in formation of intestinal air-breathing function of dojo loach, Misgurnus anguillicaudatus. Sci. Rep. 2016, 6, 31845. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lei, S.; Du, K.; Li, L.; Pang, X.; Wang, Z.; Wei, M.; Fu, S.; Hu, L.; Xu, L. Rna-seq based transcriptomic analysis uncovers α-linolenic acid and jasmonic acid biosynthesis pathways respond to cold acclimation in Camellia japonica. Sci. Rep. 2016, 6, 36463. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Wong, Y.H.; Sun, J.; He, L.S.; Chen, L.G.; Qiu, J.W.; Qian, P.Y. High-throughput transcriptome sequencing of the cold seep mussel Bathymodiolus platifrons. Sci. Rep. 2015, 5, 16597. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
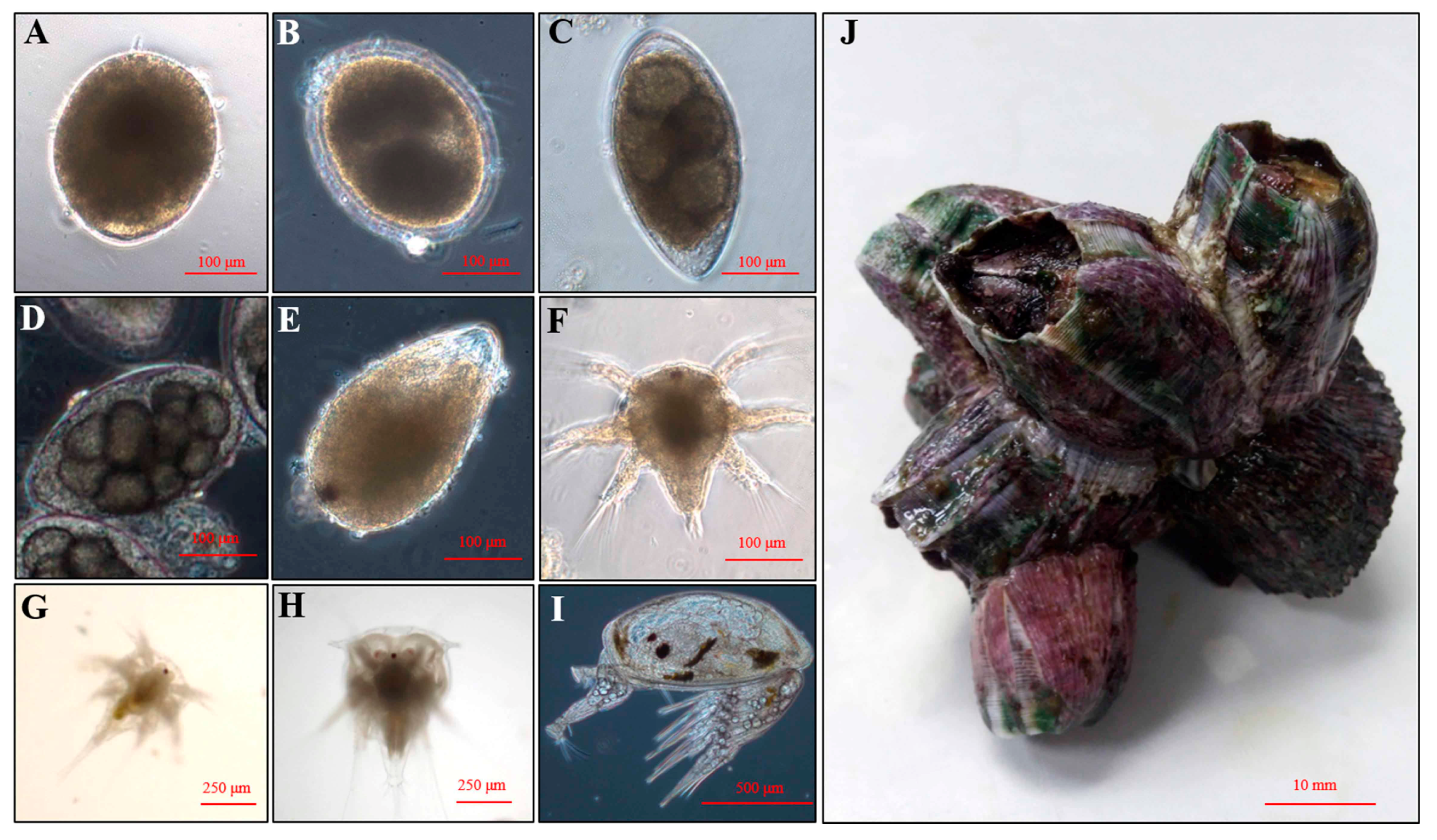

| Results | Megabalanus volcano |
|---|---|
| Output result | |
| Raw reads | 90,674,174 |
| Clean reads | 87,416,892 (96.41%) |
| Assembly result | |
| Unigene number | 42,620 |
| Unigene mean length (nt) | 865 |
| Unigene N50 (nt) | 1532 |
| Annotation result | |
| Nr | 19,522 |
| SwissProt | 15,691 |
| COG | 14,459 |
| KEGG | 10,914 |
| Gene Ontology | 8745 |
| Coding sequence prediction | |
| CDS predicted from BLAST result | 19,666 |
| CDS predicted by ESTscan | 7864 |
| Total CDS predicted | 27,530 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, G.; Zhang, G.; Huang, J.; Lan, Y.; Sun, J.; Zeng, C.; Wang, Y.; Qian, P.-Y.; He, L. Comparative Transcriptomic Analysis Reveals Candidate Genes and Pathways Involved in Larval Settlement of the Barnacle Megabalanus volcano. Int. J. Mol. Sci. 2017, 18, 2253. https://doi.org/10.3390/ijms18112253
Yan G, Zhang G, Huang J, Lan Y, Sun J, Zeng C, Wang Y, Qian P-Y, He L. Comparative Transcriptomic Analysis Reveals Candidate Genes and Pathways Involved in Larval Settlement of the Barnacle Megabalanus volcano. International Journal of Molecular Sciences. 2017; 18(11):2253. https://doi.org/10.3390/ijms18112253
Chicago/Turabian StyleYan, Guoyong, Gen Zhang, Jiaomei Huang, Yi Lan, Jin Sun, Cong Zeng, Yong Wang, Pei-Yuan Qian, and Lisheng He. 2017. "Comparative Transcriptomic Analysis Reveals Candidate Genes and Pathways Involved in Larval Settlement of the Barnacle Megabalanus volcano" International Journal of Molecular Sciences 18, no. 11: 2253. https://doi.org/10.3390/ijms18112253