Towards Better Understanding of Pea Seed Dormancy Using Laser Desorption/Ionization Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
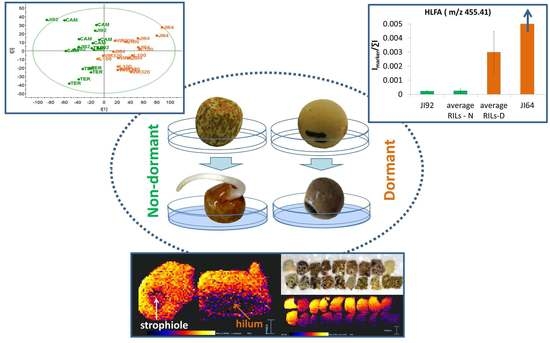
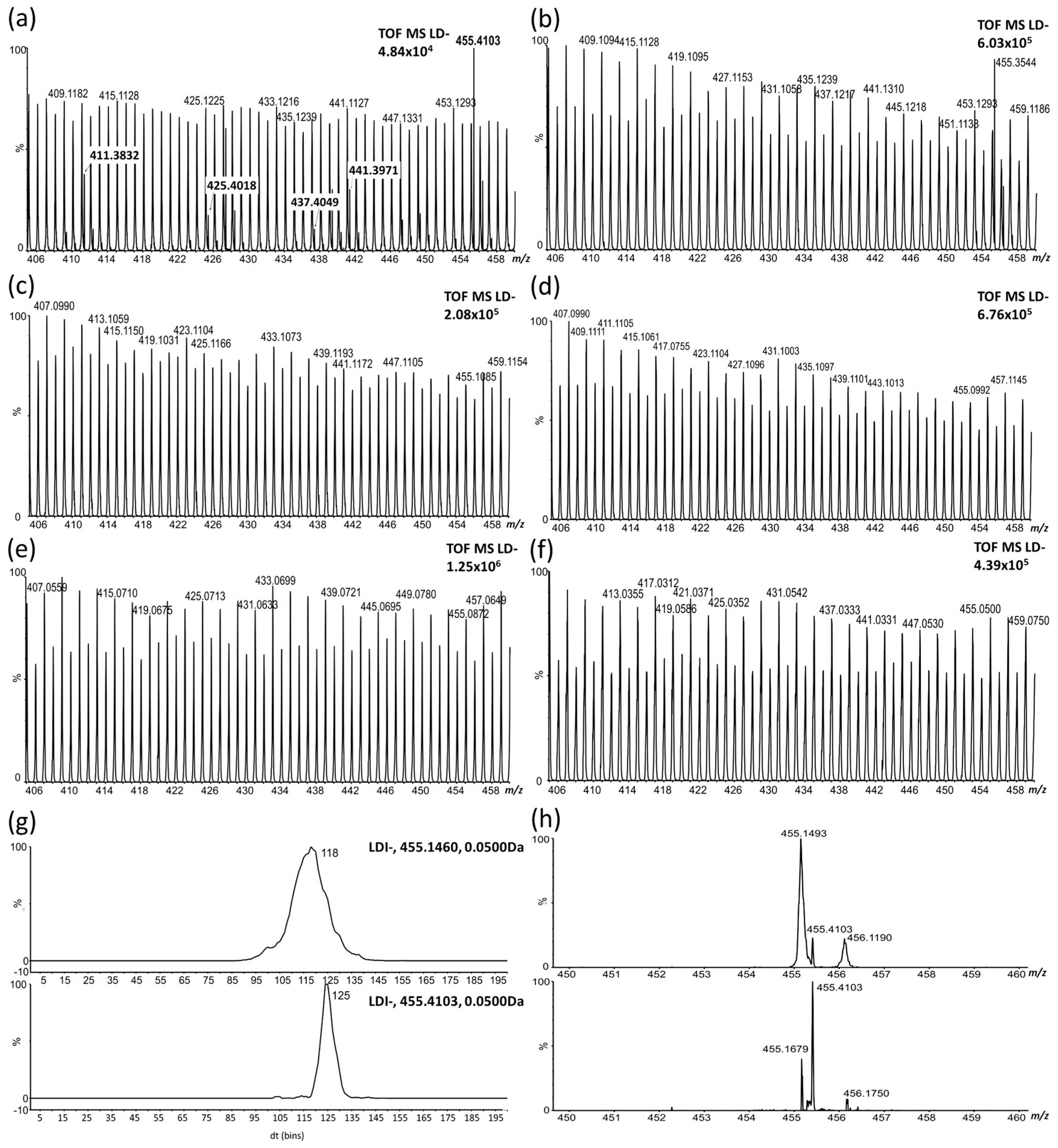
2.1. LDI-MS Measurement and Utilization of PCA and OPLS-DA for Data Analysis
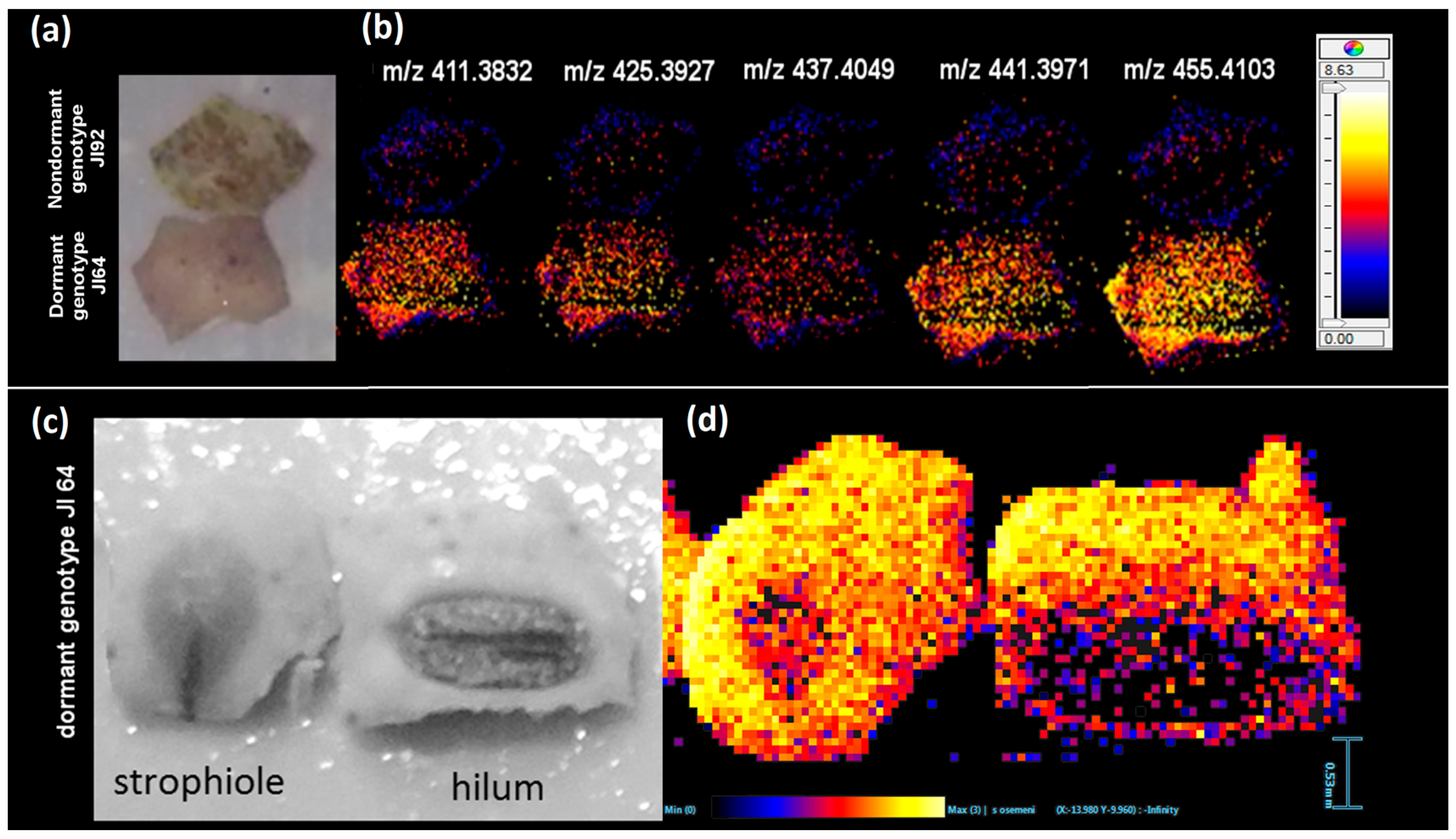
2.2. Study of the Distribution of Hydroxylated Long-Chain Fatty Acids on the Pea Seed Coat Surface
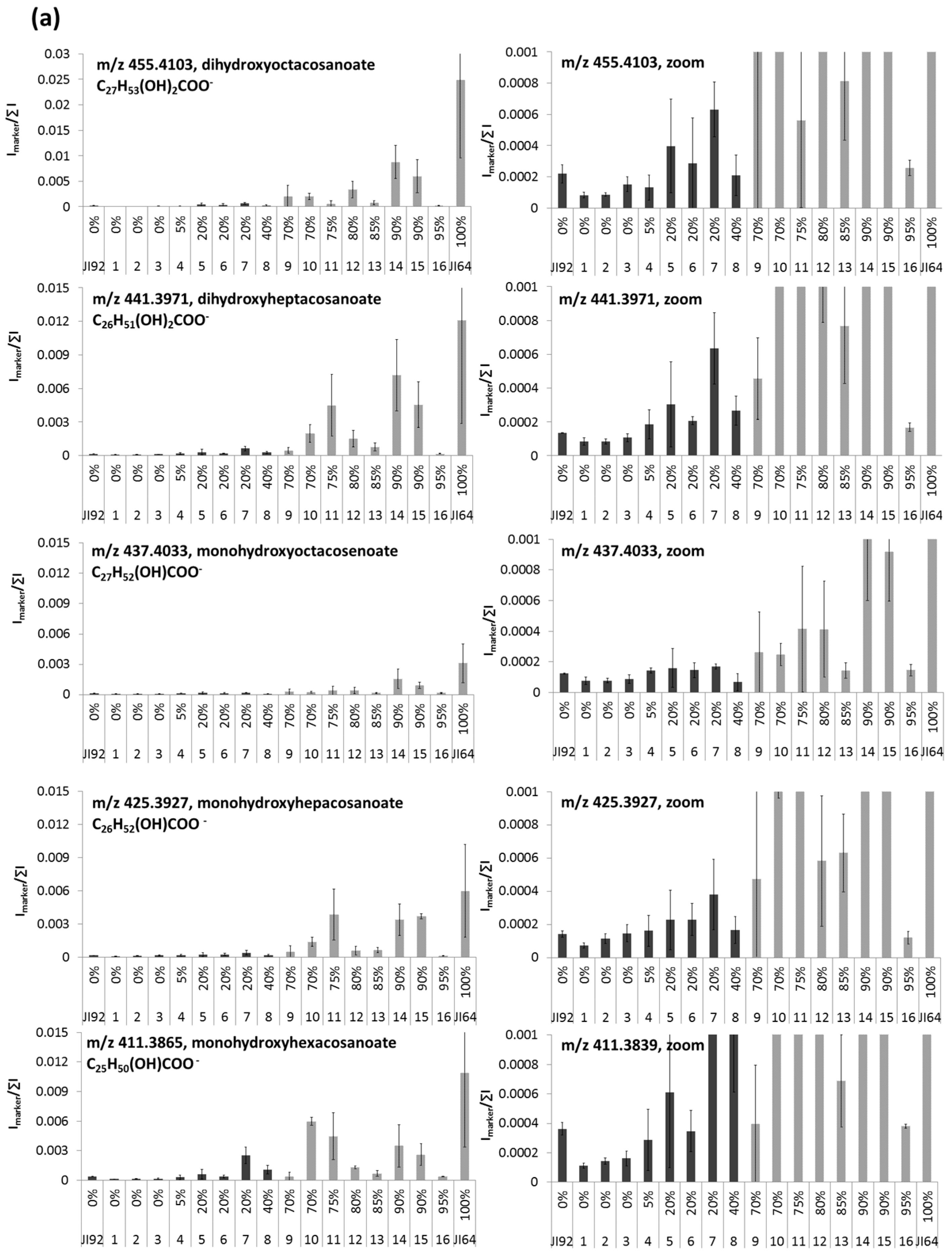
2.3. LDI-MS Analysis of HLFA in Recombinant Inbred Lines (RILs)
3. Materials and Methods
3.1. Chemicals
3.2. Plant Material
3.3. Sample Preparation for Surface Analysis
3.4. Instrumentation
Laser Desorption/Ionization Imaging Mass Spectrometry (LDI-MSI)
3.5. Data Processing
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rodríguez-Gacio, M.C.; Matilla-Vázquez, M.A.; Matilla, A.J. Seed dormancy and ABA signaling. Plant Signal. Behav. 2009, 11, 1035–1048. [Google Scholar] [CrossRef]
- Bentsink, L.; Koornneef, M. Seed Dormancy and Germination. Arabidopsis Book 2008, 6, e0119. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Vernoud, V.; Blair, M.W.; Soukup, A.; Thompson, R.D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 2014, 5, 351. [Google Scholar] [PubMed]
- Hradilová, I.; Trněný, O.; Válková, M.; Cechová, M.; Janská, A.; Prokešová, L.; Aamir, K.; Krezdorn, N.; Rotter, B.; Winter, P.; et al. A combined comparative transcriptomic, metabolomic and anatomical analyses of two key domestication traits: Pod dehiscence and seed dormancy in pea (Pisum sp.). Front. Plant Sci. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Meyer, C.J.; Ma, F.; Peterson, C.A.; Bernards, M.A. The outermost cuticle of soybean seeds: Chemical composition and function during imbibition. J. Exp. Bot. 2007, 58, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Kennedy, J.A.; Reed, B.M. Seed-coat anatomy and proanthocyanidins contribute to the dormancy of Rubus seed. Sci. Hort. 2011, 130, 762–768. [Google Scholar] [CrossRef]
- Vu, D.T.; Velusamy, V.; Park, E. Structure and chemical composition of wild soybean seed coat related to its permeability. Pak. J. Bot. 2014, 46, 1847–1857. [Google Scholar]
- Fernandez, H.; Doumas, P.; Bonnet-Masimbert, M. Quantification of GA1, GA3, GA4, GA7, GA8, GA9, GA19 and GA20; and GA20 metabolism in dormant and non-dormant beechnuts. Plant Growth Regul. 1997, 22, 29–35. [Google Scholar] [CrossRef]
- Metzger, J.D. Role of Endogenous Plant Growth Regulators in Seed Dormancy of Avena fatua: II. Gibberellins. Plant Physiol. 1983, 73, 791–795. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, D.R.; Kendall, S.L.; Florance, H.; Fedi, F.; Moore, K.; Paszkiewicz, K. Seed production temperature regulation of primary dormancy occurs through control of seed coat phenylpropanoid metabolism. New Phytol. 2015, 205, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Kanya, T.C.S.; Rao, L.J.; Sastry, M.C.S. Characterization of wax esters, free fatty alcohols and free fatty acids of crude wax from sunflower seed oil refineries. Food Chem. 2007, 101, 1552–1557. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, S.; Lee, H.M.; Lee, B.W.; Ha, T.J.; Bae, D.W.; Son, B.; Kim, Y.H.; Beak, S.; Kim, Y.C.; et al. Comparative proteomics analysis of seed coat from two black colored soybean cultivars during seed development. Plant Omnics. 2013, 6, 456–463. [Google Scholar]
- Van Malderen, S.J.M.; Laforce, B.; Van Acker, T.; Vincze, L.; Vanhaecke, F. Imaging the 3D trace metal and metalloid distribution in mature wheat and rye grains via laser ablation-ICP-mass spectrometry and micro-X-ray fluorescence spectrometry. J. Anal. At. Spectrom. 2017, 2, 289–298. [Google Scholar] [CrossRef]
- Kumar, P.M.; Srimany, A.; Ravikanth, G.; Shaanker, R.U.; Pradeep, T. Ambient ionization mass spectrometry imaging of rohitukine, a chromone anti-cancer alkaloid, during seed development in Dysoxylum binectariferum Hook.f (Meliaceae). Phytochemistry 2015, 116, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Shao, Y.; Li, A.; Zhang, Y.; Wei, C.; Wang, Y. FT-IR and NMR study of seed coat dissected from different colored progenies of Brassica napus–Sinapis alba hybrids. J. Sci. Food Agric. 2013, 93, 1898–1902. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Hua, Z.; Qian, G.; Wang, M.; Du, G.; Chen, J. Analysis of the chemical composition of cotton seed coat by Fourier-transform infrared (FT-IR) microspectroscopy. Cellulose 2009, 16, 1099–1107. [Google Scholar] [CrossRef]
- Glidewell, S. NMR imaging of developing barley grains. J. Cereal Sci. 2006, 43, 70–78. [Google Scholar] [CrossRef]
- Horn, P.J.; Korte, A.R.; Neogi, P.B.; Love, E.; Fuchs, J.; Strupat, K.; Borisjuk, L.; Shulaev, V.; Lee, Y.; Chapman, K.D. Spatial Mapping of Lipids at Cellular Resolution in Embryos of Cotton. Plant Cell 2012, 24, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Lesiak, A.D.; Cody, R.B.; Dane, A.J.; Musah, R.A. Plant Seed Species Identification from Chemical Fingerprints: A High-Throughput Application of Direct Analysis in Real Time Mass Spectrometry. Anal. Chem. 2015, 87, 8748–8757. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, K.; Signor, C.L.; Vandekerckhove, J.; Thompson, R.D.; Burstin, J. Proteomics of Medicago truncatula Seed Development Establishes the Time Frame of Diverse Metabolic Processes Related to Reserve Accumulation. Plant Physiol. 2003, 133, 664–682. [Google Scholar] [CrossRef] [PubMed]
- Dam, S.; Laursen, B.S.; Ørnfelt, J.H.; Jochimsen, B.; Stærfeldt, H.H.; Friis, C.; Nielsen, K.; Goffard, N.; Besenbacher, S.; Krusell, L.; et al. The Proteome of Seed Development in the Model Legume Lotus japonicus. Plant Physiol. 2009, 149, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- Peukert, M.; Matros, A.; Lattanzio, G.; Kaspar, S.; Abadía, J.; Mock, H.P. Spatially resolved analysis of small molecules by matrix-assisted laser desorption/ionization mass spectrometric imaging (MALDI-MSI). New Phytol. 2012, 193, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.R.; Wang, Q.; Friedt, W.; Spengler, B.; Gottwald, S.; Römpp, A. High resolution mass spectrometry imaging of plant tissues: Towards a plant metabolite atlas. Analyst 2015, 140, 7696–7709. [Google Scholar] [CrossRef] [PubMed]
- Gorzolka, K.; Kölling, J.; Nattkemper, T.W.; Niehaus, K. Spatio-Temporal Metabolite Profiling of the Barley Germination Process by MALDI MS Imaging. PLoS ONE 2016, 11, e0150208. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [PubMed]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-Based Metabolomics Data for Identification of Biochemically Interesting Compounds Using OPLS Class Models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Pirkl, A.; Meier, M.; Popkova, Y.; Letzel, M.; Schnapp, A.; Schiller, J.; Dreisewerd, K. Analysis of Free Fatty Acids by Ultraviolet Laser Desorption Ionization Mass Spectrometry Using Insect Wings as Hydrophobic Sample Substrates. Anal. Chem. 2014, 86, 10763–10771. [Google Scholar] [CrossRef] [PubMed]
- Budimir, N.; Blais, J.C.; Fournier, F.; Tabet, J.C. The use of desorption/ionization on porous silicon mass spectrometry for the detection of negative ions for fatty acids. Rapid Commun. Mass Spectrom. 2006, 20, 680–684. [Google Scholar] [CrossRef] [PubMed]
- Shroff, R.; Muck, A.; Svatoš, A. Analysis of low molecular weight acids by negative mode matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3295–3300. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, T.; Martínez, E.; Manresa, A.; Oliw, E.H. Liquid chromatography/tandem mass spectrometric analysis of 7,10-dihydroxyoctadecenoic acid, its isotopomers, and other 7,10-dihydroxy fatty acids formed by Pseudomonas aeruginosa 42A2. Rapid Commun. Mass Spectrom. 2010, 24, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Kerwin, J.L.; Torvik, J.J. Identification of Monohydroxy Fatty Acids by Electrospray Mass Spectrometry and Tandem Mass Spectrometry. Anal. Biochem. 1996, 237, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Karaki, T.; Watanabe, Y.; Kondo, T.; Koike, T. Strophiole of seeds of the black locust acts as a water gap. Plant Species Biol. 2012, 27, 226–232. [Google Scholar] [CrossRef]
- Bagheri, H.; El-Soda, M.; Kyong Kim, H.; Fritsche, S.; Jung, C.; Aarts, M.G.M. Genetic Analysis of Health-Related Secondary Metabolites in a Brassica rapa Recombinant Inbred Line Population. Int. J. Mol. Sci. 2013, 14, 15561–15577. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Zhou, C.; Molina, I.; Fu, C.; Nakashima, J.; Li, G.; Zhang, W.; Park, J.; Tang, Y.; Jiang, Q.; et al. A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. USA 2016, 113, 6997–7002. [Google Scholar] [CrossRef] [PubMed]
- Bogdanova, V.S.; Galieva, E.R.; Yadrikhinskiy, A.K.; Kosterin, O.E. Inheritance and genetic mapping of two nuclear genes involved in nuclear–cytoplasmic incompatibility in peas (Pisum sativum L.). Theor. Appl. Genet. 2012, 124, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Zablatzká, L.; Smýkal, P. Estalishment of wild pea Pisum fulvum and Pisum elatius chromosome segment subtitution lines in cultivated P. sativum genetic backround. In Proceedings of the 5th International Conference on Next Generation Genomics and Integrated Breeding for Crop Improvement, Telangana, India, 18–20 February 2015; pp. 66–67. [Google Scholar]
- North, H.; Casey, R.; Domoney, C. Inheritance and mapping of seed lipoxygenase polypeptides in Pisum. Theor. Appl. Genet. 1989, 77, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Soukup, A.; Tylová, E. Essential Methods of Plant Sample Preparation for Light Microscopy, Plant Cell Morphogenesis: Methods and Protocols. Methods Mol. Biol. 2014, 1080, 1–23. [Google Scholar] [PubMed]



| Elemental Composition | Matrix Free Experiments, Figure 3a | Matrix Assisted Experiments, Figure 3b | ||||||
|---|---|---|---|---|---|---|---|---|
| [M–H]− | dtm (ppm) * | Coordinates in S-Plot | [M–H]− | dtm (ppm) * | Coordinates in S-Plot | |||
| CoeffCS(2) | p(corr)(1) | CoeffCS(2) | p(corr)(1) | |||||
| C16H31O2− | 255.2331 | 0.78 | 0.00125 | 0.90637 | 255.2326 | −1.18 | −0.00012 | −0.09994 |
| C18H33O2− | 281.2393 | −33.07 | 0.00049 | 0.74660 | 281.2530 | 15.64 | 0.00017 | 0.70174 |
| C18H35O2− | 283.2613 | −10.24 | 0.00066 | 0.85055 | 283.2625 | −6.00 | 0.00002 | 0.04306 |
| C26H51O3− | 411.3850 | 2.67 | 0.00162 | 0.80646 | 411.3816 | −5.59 | 0.00079 | 0.92125 |
| C27H53O3− | 425.3990 | −1.18 | 0.00107 | 0.81814 | 425.3963 | −7.52 | 0.00065 | 0.96154 |
| C28H53O3− | 437.3946 | −11.20 | 0.00067 | 0.77322 | 437.3929 | −15.09 | 0.00042 | 0.91775 |
| C27H53O4− | 441.3950 | 1.36 | 0.00160 | 0.83272 | 441.3978 | 7.70 | 0.00087 | 0.96626 |
| C28H55O4− | 455.4065 | −7.69 | 0.00249 | 0.84469 | 455.4138 | 8.34 | 0.00126 | 0.91329 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cechová, M.; Válková, M.; Hradilová, I.; Janská, A.; Soukup, A.; Smýkal, P.; Bednář, P. Towards Better Understanding of Pea Seed Dormancy Using Laser Desorption/Ionization Mass Spectrometry. Int. J. Mol. Sci. 2017, 18, 2196. https://doi.org/10.3390/ijms18102196
Cechová M, Válková M, Hradilová I, Janská A, Soukup A, Smýkal P, Bednář P. Towards Better Understanding of Pea Seed Dormancy Using Laser Desorption/Ionization Mass Spectrometry. International Journal of Molecular Sciences. 2017; 18(10):2196. https://doi.org/10.3390/ijms18102196
Chicago/Turabian StyleCechová, Monika, Markéta Válková, Iveta Hradilová, Anna Janská, Aleš Soukup, Petr Smýkal, and Petr Bednář. 2017. "Towards Better Understanding of Pea Seed Dormancy Using Laser Desorption/Ionization Mass Spectrometry" International Journal of Molecular Sciences 18, no. 10: 2196. https://doi.org/10.3390/ijms18102196