Detection of Ribosomal DNA Sequence Polymorphisms in the Protist Plasmodiophora brassicae for the Identification of Geographical Isolates
Abstract
:1. Introduction
2. Results
2.1. P. brassicae DNA Isolation
2.2. Nucleotide Sequence in ITS1 and ITS2 was Conserved
2.3. Sequence Variation in the Small Subunit of the Ribosomal DNA of P. brassicae
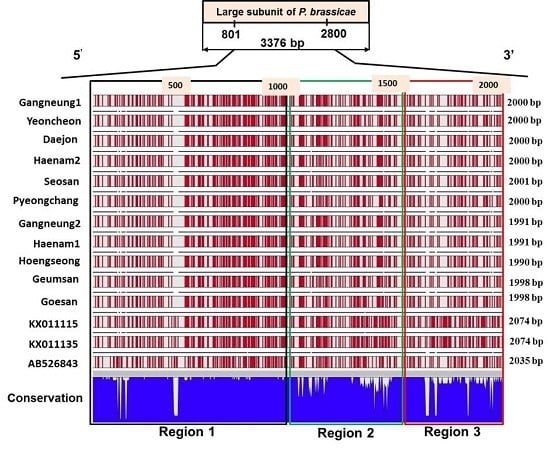
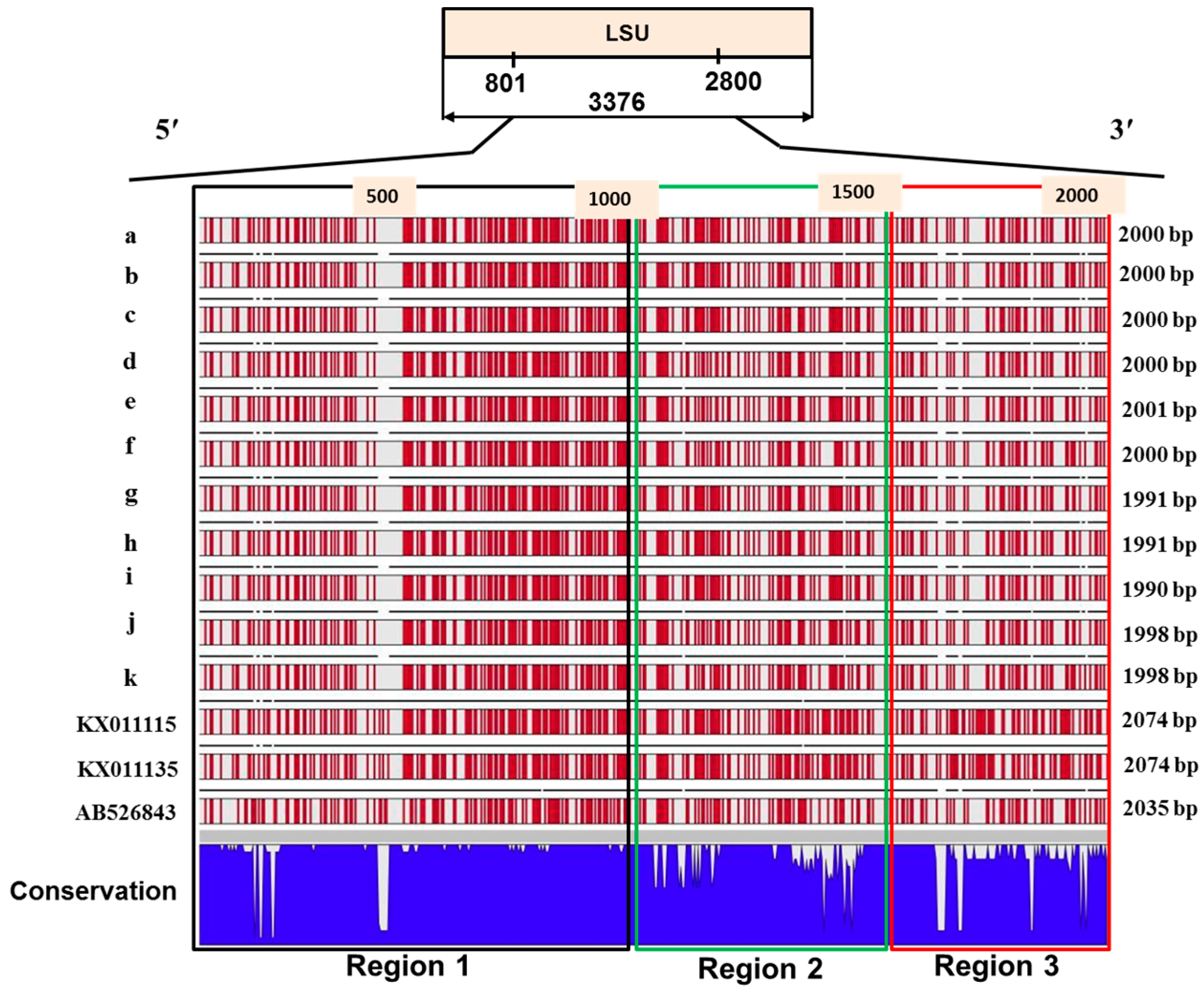
2.4. Sequence Variation in the Large Subunit of the Ribosomal DNA of P. brassicae
2.5. Variations in the Predicted Secondary Structures of the Variable Regions of the Large Subunit
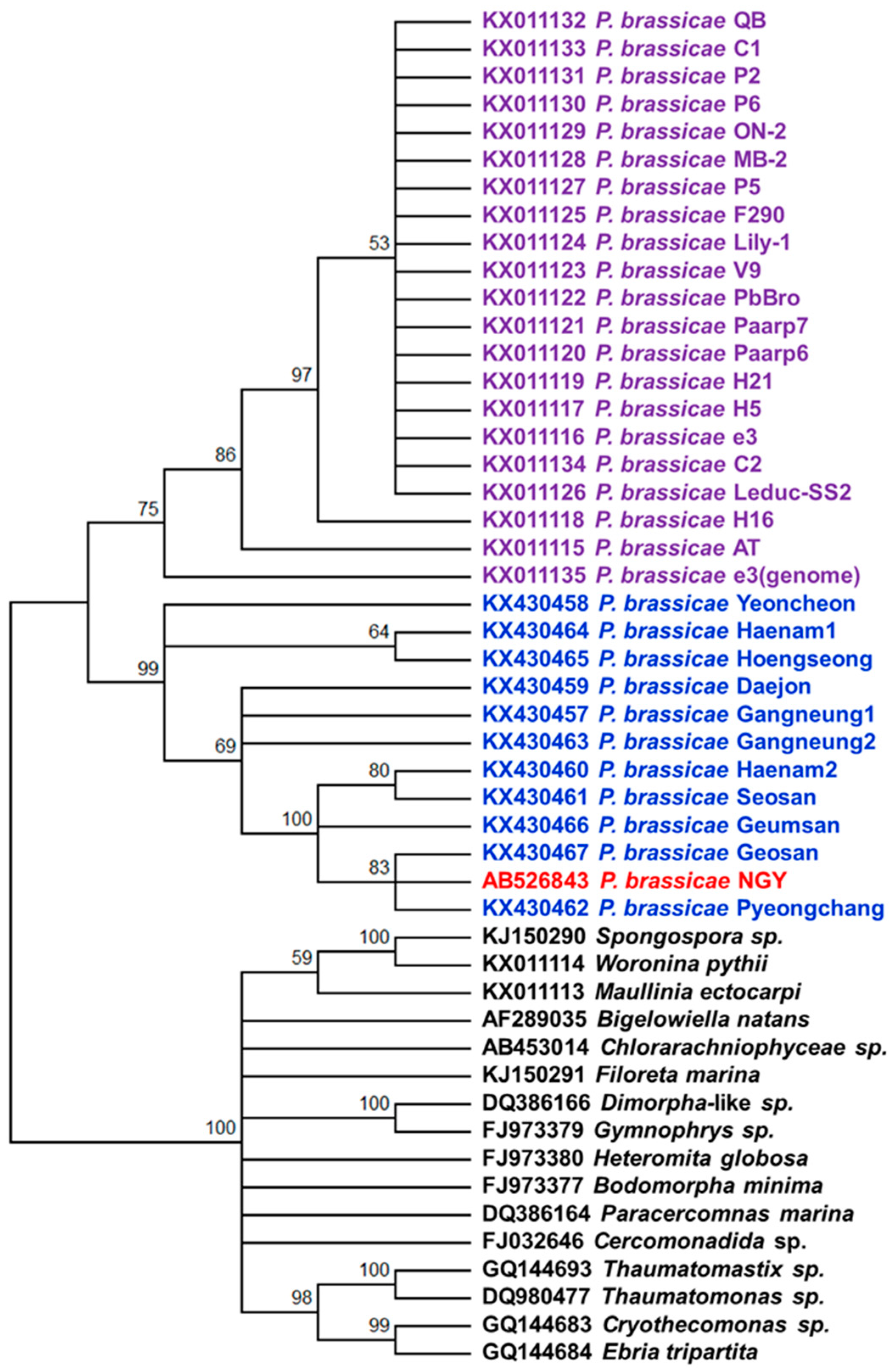
2.6. Phylogenetic Analysis
2.7. Nucleotide Polymorphisms in the Ribosomal DNA of P. brassicae
3. Discussion
3.1. Successful Isolation, Cloning and Sequencing of P. brassicae Ribosomal DNA
3.2. Ribosomal DNA Sequence Variation in the SSU and LSU of P. brassicae rDNA
3.3. Detection of Isolates Using Functional Nucleotide Variations
4. Materials and Methods
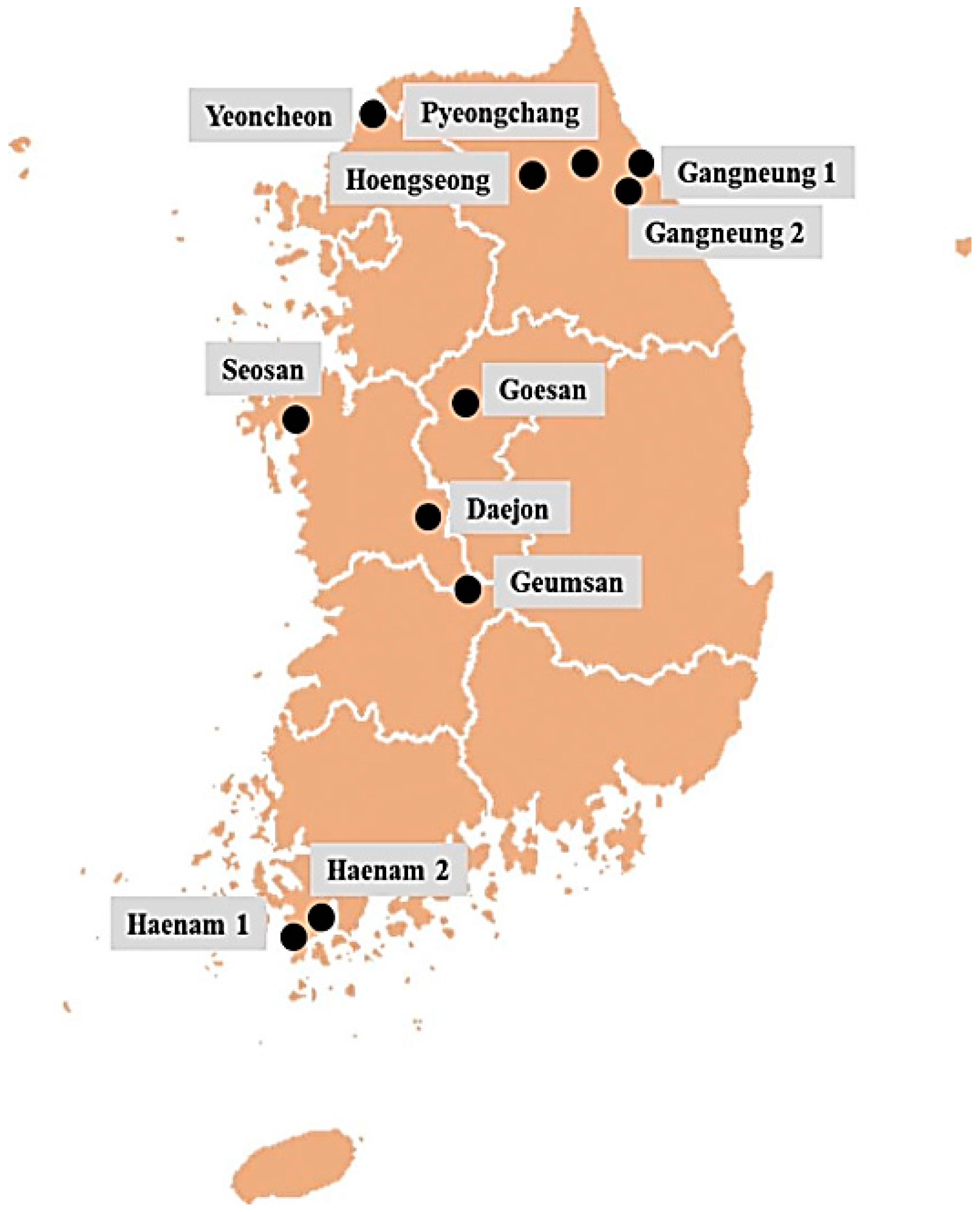
4.1. Collection of Field Populations
4.2. DNA Extraction
4.3. PCR Amplification, Cloning and Sequencing
4.4. Sequence Analysis
4.5. Probe and Primer Design and HRM Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Buczacki, S.T. Plasmodiophora. An inter-relationship between biological and practical problems. In Zoosporic Plant Pathogens: A Modern Perspective; Buczacki, S.T., Ed.; Academic Press Inc.: London, UK, 1983; pp. 161–191. [Google Scholar]
- Crete, R. Worldwide importance of clubroot. Clubroot Newslett. 1981, 11, 6–7. [Google Scholar]
- Wallenhammar, A.C. Observations on yield loss from Plasmodiophora brassicae infections in spring oilseed rape. J. Plant Dis. Prot. 1998, 105, 1–7. [Google Scholar]
- Dixon, G.R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Jing, W.; Yun, H.; Xiaoling, H.; Yingze, N.; Xiaolan, L.; Yong, L. Study of symptoms, yield loss of clubroot and modality of Plasmodiophora brassicae in rape. Chin. J. Oil Seed Sci. 2008, 30, 112–115. [Google Scholar]
- Karling, J.S. The Plasmodiophorales; Hafner Publishing: New York, NY, USA, 1968. [Google Scholar]
- Ikegami, H.; Ito, T.; Imuro, Y.; Naiki, T. Growth of Plasmodiophora brassicae in the root and callus of Chinese cabbage. In Chinese Cabbage; Talekar, N.S., Griggs, T.D., Eds.; Asian Vegetable Research and Development Center: Tainan, Taiwan, 1981; pp. 81–90. [Google Scholar]
- Ingram, D.S.; Tommerup, I.C. The life history of Plasmodiophora brassicae Woron. Proc. R. Soc. Lond. B Biol. Sci. 1972, 180, 103–112. [Google Scholar] [CrossRef]
- Naiki, T.; Dixon, G.R. The effects of chemicals on developmental stages of Plasmodiophora brassicae (clubroot). Plant Pathol. 1987, 36, 316–327. [Google Scholar] [CrossRef]
- Badi, A.M. Morphological and Molecular Studies of Diversity in Clubroot Caused by Plasmodiophora brassicae. Doctoral Dissertation, RMIT University, Melbourne, Australia, 30 August 2013. [Google Scholar]
- Voorips, E.R. Plasmodiophora brassicae: Aspects of pathogenesis and resistance in Brassica oleracea. Euphytica 1995, 83, 139–146. [Google Scholar] [CrossRef]
- Dalton, J. Effect of Plasmodiophora brassicae Resting Spore Concentration and Crop Rotation on Growth of Clubroot-Resistant Crops. Master of Science Dissertation, University of Guelph, Guelph, ON, Canada, January 2016. [Google Scholar]
- Wallenhammar, A.C. Prevalence of Plasmodiophora brassicae in spring oilseed rape growing area in Central Sweden and factors influencing soil infestation levels. Plant Pathol. 1996, 45, 710–719. [Google Scholar] [CrossRef]
- Archibald, J.M.; Keeling, P.J. Actin and ubiquitin protein sequences support a cercozoan/foraminiferan ancestry for the plasmodiophorid plant pathogens. J. Eukaryot. Microbiol. 2004, 51, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Chao, E.E.Y. Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 2003, 154, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Buczacki, S.; Toxopeus, H.; Mattusch, P.; Johnston, T.; Dixon, G.R.; Hobolth, L.A. Study of physiologic specialization in Plasmodiophora brassicae: Proposals for attempted rationalization through an international approach. Trans. Br. Mycol. Soc. 1975, 65, 295–303. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Fujimura, M.; Ishida, M.; Suzuki, T. New classification method for Plasmodiophora brassicae field isolates in Japan based on resistance of F1 cultivars of Chinese cabbage (Brassica rapa L.) to clubroot. Breed. Sci. 2004, 54, 197–201. [Google Scholar] [CrossRef]
- Kuginuki, Y.; Yoshikawa, H.; Hirai, M. Variation in virulence of Plasmodiophora brassicae in Japan tested with club root resistant cultivars of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Eur. J. Plant Pathol. 1999, 105, 327–332. [Google Scholar] [CrossRef]
- Somé, A.; Manzanares, M.J.; Laurens, F.; Baron, F.; Thomas, G.; Rouxel, F. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 1996, 45, 432–439. [Google Scholar] [CrossRef]
- Williams, P.H. A system for determination of races of Plasmodiophora brassicae that infect Cabbage and Rutabaga. Phytopathology 1966, 56, 624–626. [Google Scholar]
- Faggian, R.; Strelkov, S.E. Detection and measurement of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 282–288. [Google Scholar] [CrossRef]
- Wallenhammar, A.C.; Arwidsson, O. Detection of Plasmodiophora brassicae by PCR in naturally infested soils. Eur. J. Plant Pathol. 2001, 107, 313–321. [Google Scholar] [CrossRef]
- Ito, S.; Maehara, T.; Tanaka, S.; Kameya-Iwaki, M.; Yano, S.; Kishi, F. Cloning of a single-copy DNA sequence unique to Plasmodiophora brassicae. Physiol. Mol. Plant Pathol. 1997, 50, 289–300. [Google Scholar] [CrossRef]
- Faggian, R.; Bulman, S.R.; Lawrie, A.C.; Porter, I.J. Specific polymerase chain reaction primers for the detection of Plasmodiophora brassicae in soil and water. Phytopathology 1999, 89, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Tewari, J.; Strelkov, S.E. Molecular detection of Plasmodiophora brassicae, causal agent of clubroot of crucifers, in plant and soil. Plant Dis. 2007, 91, 80–87. [Google Scholar] [CrossRef]
- Moss, T.; Stefanovsky, V.Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase 1. Prog. Nucl. Acid Res. Mol. Biol. 1995, 50, 25–66. [Google Scholar]
- Paule, M.R.; Lofquist, A.K. Organization and expression of eukaryotic ribosomal RNA genes. In Ribosomal RNA Structure, Evolution, Processing and Function in Protein Biosynthesis; Zimmermann, R.A., Dahlberg, A.E., Eds.; CRC Press: New York, NY, USA, 1996; pp. 395–419. [Google Scholar]
- Kahl, G. Architecture of Eukaryotic Genes; VCH Verlagsgesellschaft: Weinheim, Germany, 1988. [Google Scholar]
- Poczai, P.; Hyvonen, J. Nuclear ribosomal spacer regions in plant phylogenetics: Problems and prospects. Mol. Biol. Rep. 2010, 37, 1897–1912. [Google Scholar] [CrossRef] [PubMed]
- Richard, G.F.; Kerrest, A.; Dujon, B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008, 72, 686–727. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Kawahara, A.; Murakami, H.; Tanaka, S.; Ezawa, T. Complete structure of nuclear rDNA of the obligate plant parasite Plasmodiophora brassicae: Intraspecific polymorphisms in the exon and group I intron of the large subunit rDNA. Protist 2011, 162, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Berney, C.; Dixelius, C.; Bassc, D.; Neuhauser, S. The Large Subunit rDNA Sequence of Plasmodiophora brassicae Does not Contain Intra-species Polymorphism. Protist 2016, 167, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.J.; Shim, S.A.; Jang, K.S.; Choi, Y.H.; Kim, J.C.; Choi, G.J. Resistance of cultivars of Chinese cabbage to Plasmodiophora brassicae isolates of several races collected in Korea. Korean J. Hort. Sci. Technol. 2011, 29, 610–616. [Google Scholar]
- Ward, E.; Adams, M.J. Analysis of ribosomal DNA sequences of Polymyxa species and related fungi and the development of genus-and species-specific PCR primers. Mycol. Res. 1998, 102, 965–974. [Google Scholar] [CrossRef]
- Wallenhammar, A.C.; Almquist, C.; Söderström, M.; Jonsson, A. In-field distribution of Plasmodiophora brassicae measured using quantitative real-time PCR. Plant Pathol. 2012, 61, 16–28. [Google Scholar] [CrossRef]
- Feng, J.; Hwang, S.F.; Strelkov, S.E. Assessment of gene expression profiles in primary and secondary zoospores of Plasmodiophora brassicae by dot blot and real-time PCR. Microbiol. Res. 2013, 168, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Castlebury, L.A.; Domier, L.L. Small subunit ribosomal RNA gene phylogeny of Plasmodiophora brassicae. Mycologia 1998, 90, 102–107. [Google Scholar] [CrossRef]
- Waminal, N.E.; Perumal, S.; Lim, K.B.; Park, B.S.; Kim, H.H.; Yang, T.J. Genomic Survey of the Hidden Components of the B. rapa Genome. In The Brassica rapa Genome; Wang, X., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 83–96. [Google Scholar]
- Thiéry, O.; Vasar, M.; Jairus, T.; Davison, J.; Roux, C.; Kivistik, P.A.; Zobel, M. Sequence variation in nuclear ribosomal small subunit, internal transcribed spacer and large subunit regions of Rhizophagus irregularis and Gigaspora margarita is high and isolate-dependent. Mol. Ecol. 2016, 25, 2816–2832. [Google Scholar]
- Kim, H.; Jo, E.J.; Choi, Y.H.; Jang, K.S.; Choi, G.J. Pathotype Classification of Plasmodiophora brassicae Isolates Using Clubroot-Resistant Cultivars of Chinese Cabbage. Plant Pathol. J. 2016, 32, 423. [Google Scholar] [CrossRef] [PubMed]
- Kohout, P.; Sudová, R.; Janoušková, M.; Čtvrtlíková, M.; Hejda, M.; Pánková, H.; Sýkorová, Z. Comparison of commonly used primer sets for evaluating arbuscular mycorrhizal fungal communities: Is there a universal solution. Soil Biol. Biochem. 2014, 68, 482–493. [Google Scholar] [CrossRef]
- Opik, M.; Davison, J.; Moora, M.; Zobel, M. DNA-based detection and identification of Glomeromycota: The virtual taxonomy of environmental sequences. Botany-Botanique 2014, 92, 135–147. [Google Scholar] [CrossRef]
- Hart, M.M.; Aleklett, K.; Chagnon, P.L.; Egan, C.; Ghignone, S.; Helgason, T.; Lekberg, Y.; Öpik, M.; Pickles, B.J.; Waller, L. Navigating the labyrinth: A guide to sequence-based, community ecology of arbuscular mycorrhizal fungi. New Phytol. 2015, 207, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Jo, E.J.; Jang, K.S.; Choi, Y.H.; Ahn, K.G.; Choi, G.J. Resistance of Cabbage Plants to Isolates of Plasmodiophora brassicae. Korean J. Hortic. Sci. Technol. 2016, 34, 442–452. [Google Scholar]
- Buhariwalla, H.; Mithen, R. Cloning of a Brassica repetitive DNA element from resting spores of Plasmodiophora brassicae. Physiol. Mol. Plant Pathol. 1995, 47, 95–101. [Google Scholar] [CrossRef]
- Buhariwalla, H.; Greaves, S.; Magrath, R.; Mithen, R. Development of specific PCR primers for the amplification of polymorphic DNA from the obligate root pathogen Plasmodiophora brassicae. Physiol. Mol. Plant Pathol. 1995, 47, 83–94. [Google Scholar] [CrossRef]
- Manzanares-Dauleux, M.J.; Divaret, I.; Baron, F.; Thomas, G. Assessment of biological and molecular variability between and within field isolates of Plasmodiophora brassicae. Plant Pathol. 2001, 50, 165–173. [Google Scholar] [CrossRef]
- Bryngelsson, T.; Gustafsson, M.; Green, B.; Lind, C. Uptake of host DNA by the parasitic fungus Plasmodiophora brassicae. Physiol. Mol. Plant Pathol. 1998, 33, 163–171. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]




| Name | Primer Forward (F) and Reverse (R) | Position | Product Size |
|---|---|---|---|
| CR_rDNA 1 | F: GGTTGATCCTGCCAGTAGTC | 6–774 | 768 |
| R: GAAGCGATTCGGGCATAGAG | |||
| CR_rDNA 2 | F: CCTATGCTAATCTCGTGGCG | 728–2182 | 1454 |
| R: TAAGAAGTCACGGACCTAGG | |||
| CR_rDNA 3 | F: TTATGCCTATGCCTTCCCGG | 2164–3192 | 1028 |
| R: GAAACACGCAGCTGGAGTCGC | |||
| CR_rDNA 4 | F: TCCGTAGGTGAACCTGCGGA | 3080–4142 | 1062 |
| R: CCCATTCTGACCTAGGCCAA | |||
| CR_rDNA 5 | F: TTGGCCTAGGTCAGAATGGG | 4110–5606 | 1496 |
| R: ATGCGGTTATGAGTACGACCA | |||
| CR_rDNA 6 | F: CAGGTTCATATTCCTGAACC | 5346–6981 | 1635 |
| R: CGTTCAAATCAAGTCGTCTAC |
| Different Geographic Isolates | Accession No. | Sequence Length (bp) | Sequence Encoding |
|---|---|---|---|
| Gangneung1 | KX430457 | 6931 | Partial ribosomal RNA, both small and large subunits |
| Yeoncheon | KX430458 | 6937 | |
| Daejon | KX430459 | 6935 | |
| Haenam2 | KX430460 | 6927 | |
| Seosan | KX430461 | 6923 | |
| Pyeongchang | KX430462 | 6932 | |
| Gangneung2 | KX430463 | 6931 | |
| Haenam1 | KX430464 | 6935 | |
| Hoengseong | KX430465 | 6934 | |
| Geumsan | KX430466 | 6923 | |
| Goesan | KX430467 | 6929 |
| Probe Name | Position | Probe Sequence | Nucleotide Variation | Froward (F) and Reverse (R) Primers | Position | Product Size (bp) |
|---|---|---|---|---|---|---|
| Probe_594 | 594 | CAGTGTCTGTTTACTGTTGGCGCCA | A/G | F: TTAAACCTATTATCGAGGATCC R: ACGAGATTAGCATAGGATTATC | 530–742 | 212 |
| Probe_629 | 629–632 | GGCTTCTCTTTT---AAAGAAGTTATGC | ---/ATT | |||
| Probe_661 | 662 | TCGAGCAGCCCATTCTAGTTGTGGG | T/A | |||
| Probe_856 | 857 | TCGAAAGAATGCCGATCGACTGGAG | C/T | F: GAAGCGATTCGGGCATAGAGGCC R: CGCTATTGGAGCTGGTATTACC | 756–982 | 226 |
| Probe_903 | 904–912 | CGGGGGTGTGTG----TTTGATGGAGGAA | ----/CGCG | |||
| Probe_1364 | 1366 | AGCATTCACCAA-GGATGTCCTCTTT | -/A | F: GAAGTAATGATTGATAGGGATAGT R: CGACCGCCAATCCCTAGTCGAC | 1262–1465 | 203 |
| Probe_3564 | 3561–3562 | ACACACACACACA_TCAAAGATCTCA | --/CA | F: GCTGCGGCATAGCTTGAACGAA R: CGGTTTGAGCTCTTCCCGCTTC | 3448–3685 | 237 |
| Probe_5516 | 5545 | TCCGACGGACAGACTTATTCGCA | C/G, C/A, C/T | F: TCAGGTTCATATTCCTGAACCA R: ACCTGATGCGGTTATGAGTACG | 5316–5574 | 258 |
| Probe_6479 | 6479 | ATGAATACAAACTGTGAAAGCATGGC | T/C | F: CGATCTACGCGAGAGACAAAGTC R: GCAACGTCGCTATGAACGCTTG | 6315–6594 | 279 |
| Probe_6759 | 6759 | AGTAATCCAATTCAGTACGAGAGGA | C/T | F: ATCATTGCCTCGCAGCAGAGGCC R: AGAGGCGTTCAGCCGTAATCCT | 6624–6860 | 236 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laila, R.; Robin, A.H.K.; Yang, K.; Choi, G.J.; Park, J.-I.; Nou, I.-S. Detection of Ribosomal DNA Sequence Polymorphisms in the Protist Plasmodiophora brassicae for the Identification of Geographical Isolates. Int. J. Mol. Sci. 2017, 18, 84. https://doi.org/10.3390/ijms18010084
Laila R, Robin AHK, Yang K, Choi GJ, Park J-I, Nou I-S. Detection of Ribosomal DNA Sequence Polymorphisms in the Protist Plasmodiophora brassicae for the Identification of Geographical Isolates. International Journal of Molecular Sciences. 2017; 18(1):84. https://doi.org/10.3390/ijms18010084
Chicago/Turabian StyleLaila, Rawnak, Arif Hasan Khan Robin, Kiwoung Yang, Gyung Ja Choi, Jong-In Park, and Ill-Sup Nou. 2017. "Detection of Ribosomal DNA Sequence Polymorphisms in the Protist Plasmodiophora brassicae for the Identification of Geographical Isolates" International Journal of Molecular Sciences 18, no. 1: 84. https://doi.org/10.3390/ijms18010084