Identification of the MUC2 Promoter as a Strong Promoter for Intestinal Gene Expression through Generation of Transgenic Quail Expressing GFP in Gut Epithelial Cells
Abstract
:1. Introduction
2. Results
2.1. Microarray Analysis of Intestine-Specific Mucin Genes in Mouse and Human
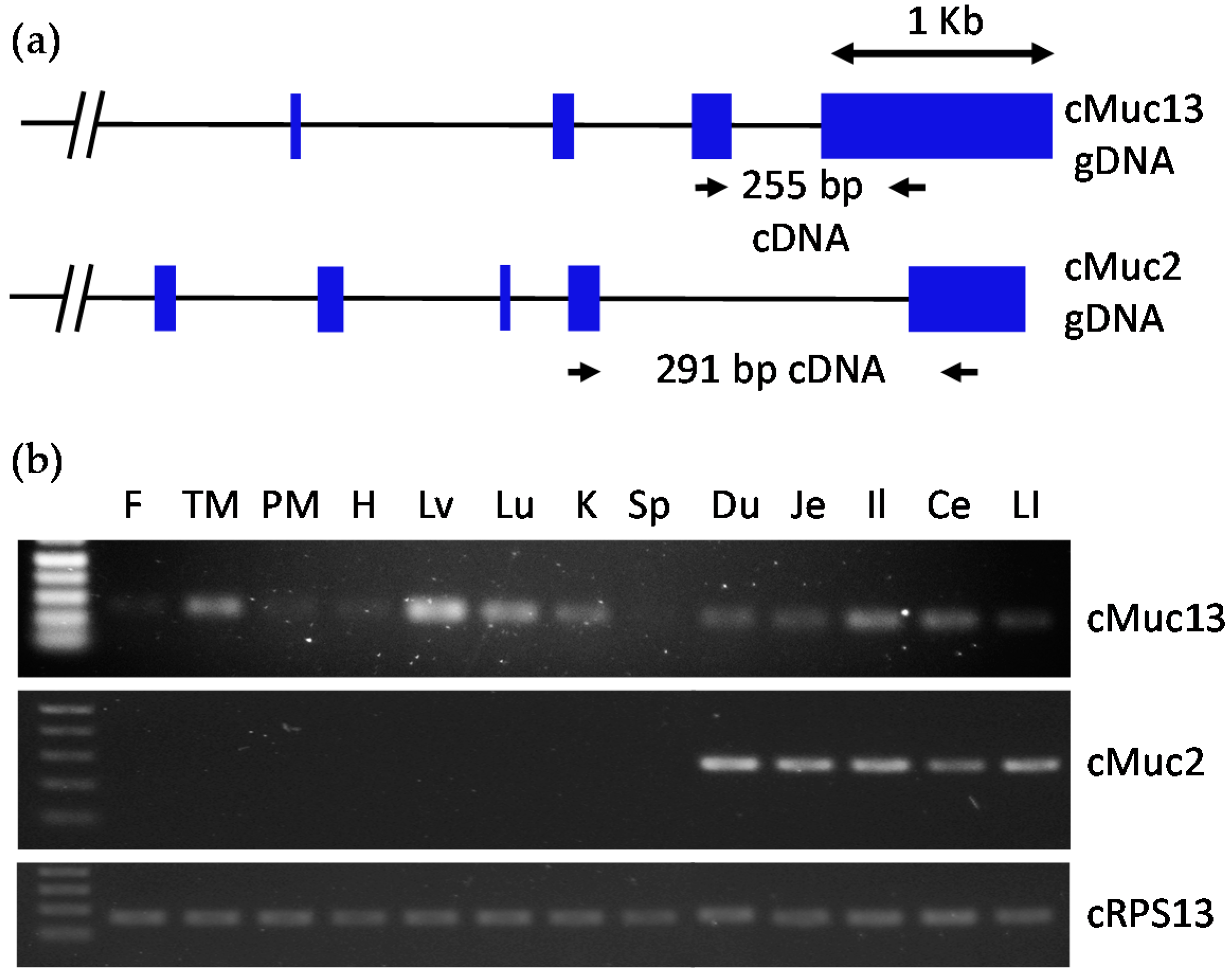
2.2. Confirmation of Intestine-Specific Expression of Mucin Genes in Chickens
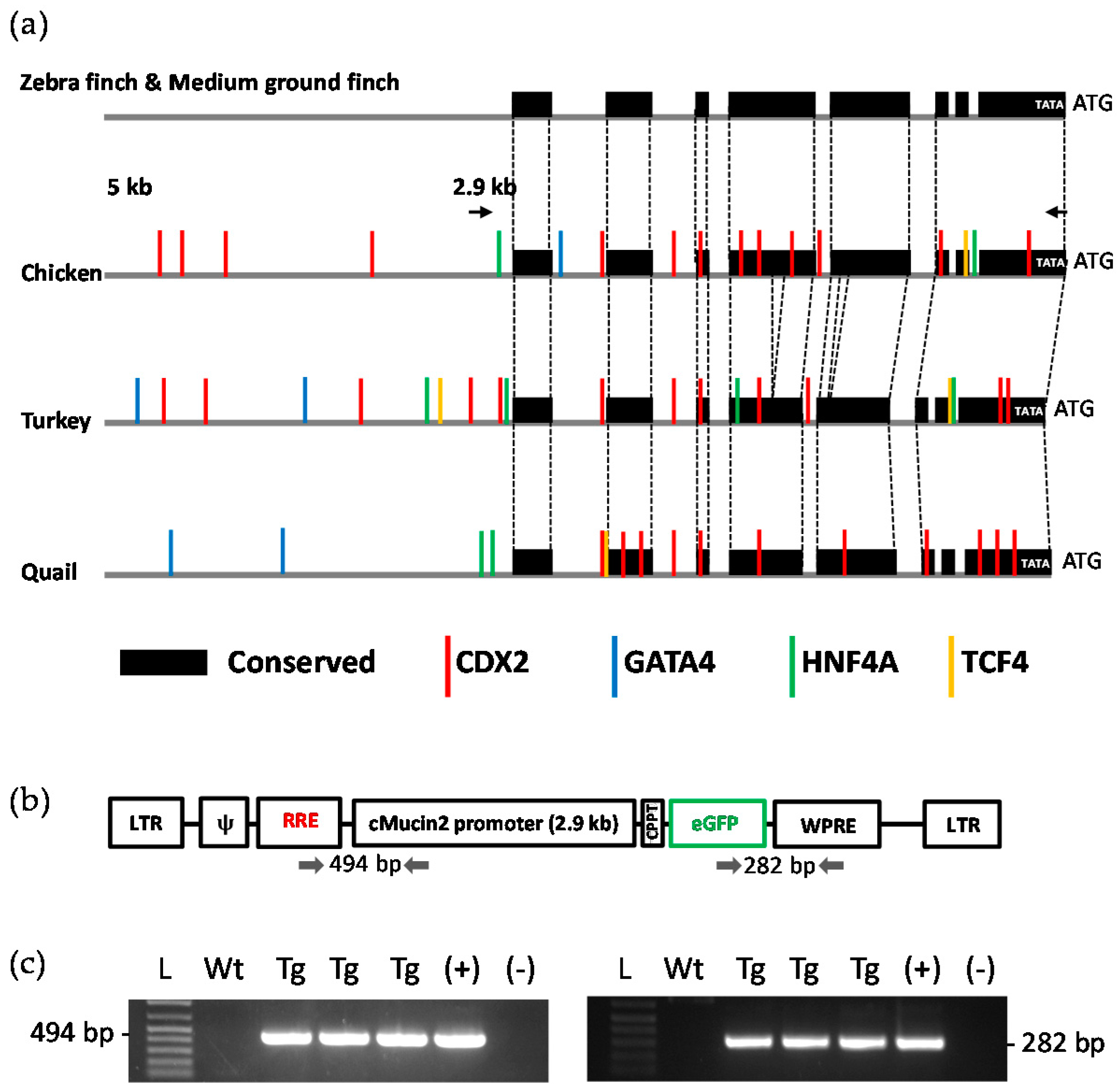
2.3. Analysis of the MUC2 Promoter
2.4. Generation of Transgenic Birds
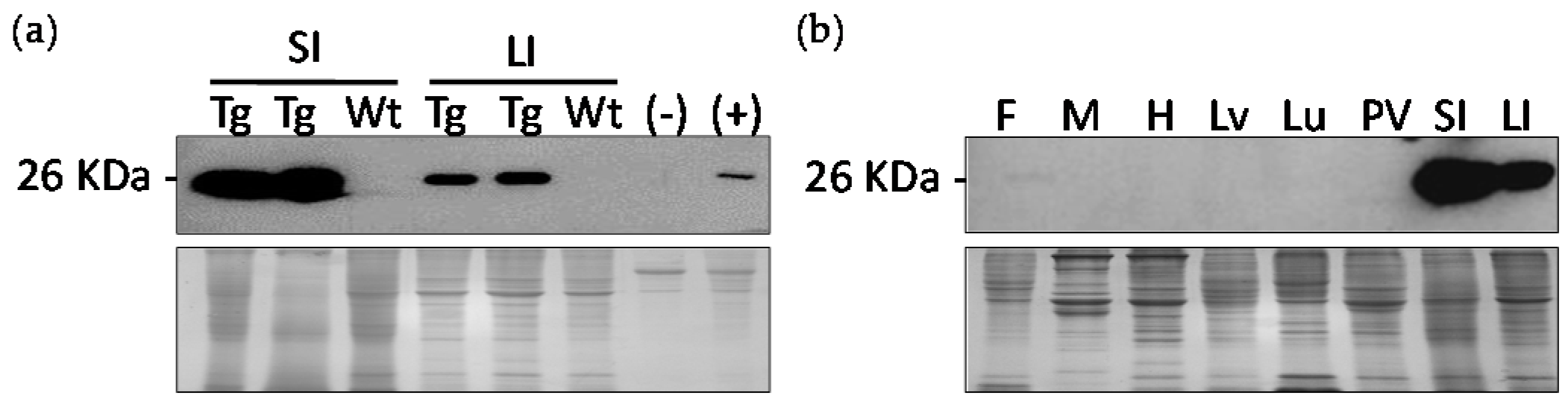
2.5. Western Blot Analysis of Transgenic Birds for Tissue Distribution
2.6. Protein and Fluorescence Detection of eGFP in Epithelial Layer of Intestinal Tissues
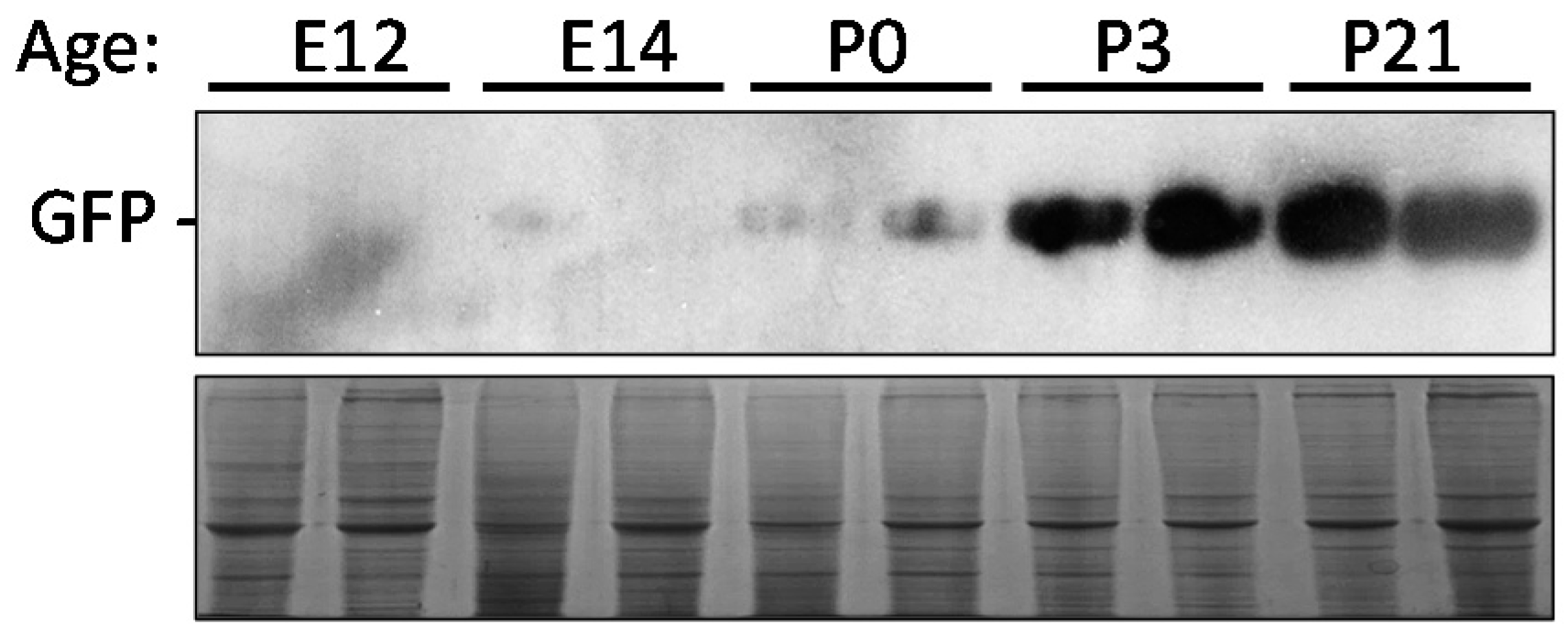
2.7. Time Point Expression of eGFP
3. Discussion
4. Materials and Methods
4.1. Animal Use and Ethics Statement
4.2. Data Mining Using GEO DataSets
4.3. Analysis of Transcription Factor Binding Sites on the Promoter Region
4.4. Total RNA Extraction, cDNA Synthesis, and PCR
4.5. Vector Construction and Production of Lentiviral Particles
4.6. Production of the Founder Quail
4.7. Mating and Selection of Transgenic Offspring
4.8. Tissue Collection and Microscopic Examination of GFP Expression
4.9. Western Blot Analysis
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MUC2 | Mucin 2 |
| GFP | Green fluorescence protein |
| CDX2 | Caudal type homeobox 2 |
| GATA4 | GATA binding protein 4 |
| HNF4A | Hepatocyte nuclear factor 4 alpha |
| TCF4 | Transcription factor 4 |
| aFABP | Adipocyte fatty acid binding protein |
| I-FABP | Intestinal fatty acid binding protein |
| GEO | Gene Expression Omnibus |
| GDS | GEO DataSet |
References
- Song, Y.; Ahn, J.; Suh, Y.; Davis, M.E.; Lee, K. Identification of novel tissue-specific genes by analysis of microarray databases: A human and mouse model. PLoS ONE 2013, 8, e64483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ahn, J.; Suh, Y.; Hwang, S.; Davis, M.E.; Lee, K. Identification of CTLA2A, DEFB29, WFDC15B, SERPINA1F and MUP19 as novel tissue-specific secretory factors in mouse. PLoS ONE 2015, 10, e0124962. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Li, B.; Davis, M.E.; Suh, Y.; Lee, K. Comparative analysis of fatty acid-binding protein 4 promoters: Conservation of peroxisome proliferator-activated receptor binding sites. J. Anim. Sci. 2009, 87, 3923–3934. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Shin, S.; Suh, Y.; Park, J.Y.; Hwang, S.; Lee, K. Identification of the avian RBP7 gene as a new adipose-specific gene and RBP7 promoter-driven GFP expression in adipose tissue of transgenic quail. PLoS ONE 2015, 10, e0124768. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Ahn, J.; Suh, Y.; Moeller, S.J.; Hwang, S.; Lee, K. Isolation and in vitro validation of cardiac muscle-specific promoters in pigs. Cell. Mol. Biol. 2016. [Google Scholar] [CrossRef]
- Lee, K.; Villena, J.A.; Moon, Y.S.; Kim, K.H.; Lee, S.; Kang, C.; Sul, H.S. Inhibition of adipogenesis and development of glucose intolerance by soluble Pref-1. J. Clin. Investig. 2003, 111, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lee, K.; Moon, Y.S.; Ahmadian, M.; Kim, K.H.; Roder, K.; Kang, C.; Sul, H.S. Overexpression of pref-1 in pancreatic islet β-cells in mice causes hyperinsulinemia with increased islet mass and insulin secretion. Biochem. Biophys. Res. Commun. 2015, 461, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Madison, B.B.; Dunbar, L.; Qiao, X.T.; Braunstein, K.; Braunstein, E.; Gumucio, D.L. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002, 277, 33275–33283. [Google Scholar] [CrossRef] [PubMed]
- Janssen, K.P.; EL-Marjou, F.; Pinto, D.; Sastre, X.; Rouillard, D.; Fouquet, C.; Soussi, T.; Louvard, D.; Robine, S. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology 2002, 123, 492–504. [Google Scholar] [CrossRef] [PubMed]
- El Marjou, F.; Janssen, K.P.; Chang, B.H.; Li, M.; Hindie, V.; Chan, L.; Louvard, D.; Chambon, P.; Metzger, D.; Robine, S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 2004, 39, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Nandan, M.O.; McConnell, B.B.; Ghaleb, A.M.; Bialkowska, A.B.; Sheng, H.; Shao, J.; Babbin, B.A.; Robine, S.; Yang, V.W. Krüppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology 2008, 134, 120–130. [Google Scholar] [CrossRef] [PubMed]
- McConnell, B.B.; Kim, S.S.; Yu, K.; Ghaleb, A.M.; Takeda, N.; Manabe, I.; Nusrat, A.; Nagai, R.; Yang, V.W. Krüppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology 2011, 141, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Sweetser, D.A.; Hauft, S.M.; Hoppe, P.C.; Birkenmeier, E.H.; Gordon, J.I. Transgenic mice containing intestinal fatty acid-binding protein-human growth hormone fusion genes exhibit correct regional and cell-specific expression of the reporter gene in their small intestine. Proc. Natl. Acad. Sci. USA 1988, 85, 9611–9615. [Google Scholar] [CrossRef] [PubMed]
- Stoltz, D.A.; Rokhlina, T.; Ernst, S.E.; Pezzulo, A.A.; Ostedgaard, L.S.; Karp, P.H.; Samuel, M.S.; Welsh, M.J. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J. Clin. Investig. 2013, 123, 2685–2693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousseau, K.; Byrne, C.; Kim, Y.S.; Gum, J.R.; Swallow, D.M.; Toribara, N.W. The complete genomic organization of the human MUC6 and MUC2 mucin genes. Genomics 2004, 83, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Voynow, J.A.; Rubin, B.K. Mucins, mucus, and sputum. Chest 2009, 135, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Longman, R.J.; Douthwaite, J.; Sylvester, P.A.; Thomas, M.G.; Poulsom, R.; Wright, N.A.; Corfield, A.P.; Thomas, M.G.; Wright, N.A. Coordinated localisation of mucins and trefoil peptides in the ulcer associated cell lineage and the gastrointestinal mucosa. Gut 2000, 47, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Piñeiro, A.M.; Bergström, J.H.; Ermund, A.; Gustafsson, J.K.; Schütte, A.; Johansson, M.E.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G348–G356. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Choi, Y.M.; Han, J.Y.; Lee, K. Inhibition of lipolysis in the novel transgenic quail model overexpressing G0/G1 switch gene 2 in the adipose tissue during feed restriction. PLoS ONE 2014, 9, e100905. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.R.; Shin, S.; Choi, Y.M.; Kim, E.; Han, J.Y.; Lee, K. Overexpression of G0/G1 switch gene 2 in adipose tissue of transgenic quail inhibits lipolysis associated with egg laying. Int. J. Mol. Sci. 2016, 17, 384. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.S.; Kim, T.M.; Kim, S.Y.; Kim, T.W.; Seo, H.W.; Lee, S.K.; Kwon, S.C.; Lee, G.S.; Kim, H.; Lim, J.M.; Han, J.Y. Generation of transgenic quail through germ cell-mediated germline transmission. FASEB J. 2008, 22, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; White, P.; Kaestner, K.H. Establishment of intestinal identity and epithelial-mesenchymal signaling by CDX2. Dev. Cell 2009, 16, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Verzi, M.P.; Shin, H.; San Roman, A.K.; Liu, X.S.; Shivdasani, R.A. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 2013, 33, 281–292. [Google Scholar] [CrossRef] [PubMed]
- San Roman, A.K.; Aronson, B.E.; Krasinski, S.D.; Shivdasani, R.A.; Verzi, M.P. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J. Biol. Chem. 2015, 290, 1850–1860. [Google Scholar] [CrossRef] [PubMed]
- Verzi, M.P.; Hatzis, P.; Sulahian, R.; Philips, J.; Schuijers, J.; Shin, H.; Freed, E.; Lynch, J.P.; Dang, D.T.; Brown, M.; et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc. Natl. Acad. Sci. USA 2010, 107, 15157–15162. [Google Scholar] [CrossRef] [PubMed]
- Lois, C.; Hong, E.J.; Pease, S.; Brown, E.J.; Baltimore, D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 2012, 295, 868–872. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.B.; Lois, C. Generation of tissue-specific transgenic birds with lentiviral vectors. Proc. Natl. Acad. Sci. USA 2005, 102, 16443–16447. [Google Scholar] [CrossRef] [PubMed]
- Lillico, S.G.; Sherman, A.; McGrew, M.J.; Robertson, C.D.; Smith, J.; Haslam, C.; Barnard, P.; Radcliffe, P.A.; Mitrophanous, K.A.; Elliot, E.A.; et al. Oviduct-specific expression of two therapeutic proteins in transgenic hens. Proc. Natl. Acad. Sci. USA 2007, 104, 1771–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Applegate, T.J.; Lossie, A.C. Cloning, annotation and developmental expression of the chicken intestinal MUC2 gene. PLoS ONE 2013, 8, e53781. [Google Scholar] [CrossRef] [PubMed]

| Gene | Fold a | S. Intestine | Spleen | Muscle | Liver | Brain | Lung | Kidney | Heart | p Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | ||||||||||
| Muc13 | 61.2 | 8463 ± 448 | 131 ± 2 | 145 ± 5 | 144 ± 7 | 124 ± 9 | 130 ± 8 | 137 ± 3 | 157 ± 6 | <0.0001 |
| Muc3 | 41.5 | 3752 ± 1093 | 82 ± 8 | 91 ± 4 | 98 ± 1 | 77 ± 3 | 82 ± 2 | 86 ± 3 | 117 ± 6 | <0.0001 |
| Muc2 | 26.9 | 2570 ± 195 | 90 ± 3 | 93 ± 2 | 100 ± 6 | 85 ± 1 | 86 ± 3 | 95 ± 5 | 120 ± 6 | <0.0001 |
| Muc4 | 5.9 | 472 ± 40 | 70 ± 3 | 69 ± 4 | 73 ± 5 | 71 ± 1 | 139 ± 11 | 68 ± 1 | 71 ± 3 | <0.0001 |
| Muc5ac | 1.2 | 157 ± 9 | 114 ± 4 | 130 ± 3 | 142 ± 5 | 113 ± 2 | 117 ± 5 | 111 ± 3 | 165 ± 3 | <0.0001 |
| Mucl1 | 1.1 | 77 ± 7 | 60 ± 3 | 78 ± 2 | 66 ± 2 | 63 ± 1 | 66 ± 5 | 66 ± 1 | 73 ± 4 | <0.05 |
| Muc5b | 1.1 | 158 ± 3 | 147 ± 2 | 119 ± 3 | 153 ± 13 | 104 ± 2 | 243 ± 21 | 132 ± 1 | 141 ± 7 | >0.05 |
| Muc16 | 1.0 | 95 ± 2 | 74 ± 6 | 77 ± 1 | 96 ± 6 | 73 ± 4 | 124 ± 7 | 80 ± 2 | 114 ± 2 | >0.05 |
| Muc20 | 1.0 | 109 ± 6 | 108 ± 6 | 106 ± 7 | 107 ± 7 | 105 ± 7 | 95 ± 3 | 104 ± 8 | 125 ± 4 | >0.05 |
| Muc15 | 1.0 | 71 ± 5 | 63 ± 1 | 69 ± 3 | 75 ± 6 | 71 ± 3 | 67 ± 2 | 68 ± 2 | 76 ± 3 | >0.05 |
| Muc1 | 0.6 | 144 ± 4 | 98 ± 6 | 102 ± 7 | 102 ± 4 | 85 ± 2 | 916 ± 33 | 198 ± 23 | 127 ± 4 | NA |
| Human | ||||||||||
| Muc13 | 50.3 | 100,638 ± 785 | 204 ± 10 | 250 ± 12 | 663 ± 172 | 277 ± 38 | 927 ± 659 | 11,295 ± 945 | 402 ± 128 | <0.0001 |
| Muc2 | 31.7 | 26,029 ± 501 | 339 ± 35 | 1168 ± 318 | 1498 ± 852 | 891 ± 324 | 694 ± 186 | 613 ± 24 | 550 ± 45 | <0.0001 |
| Muc20 | 2.0 | 50,305 ± 651 | 7068 ± 83 | 9927 ± 289 | 24,134 ± 413 | 2257 ± 260 | 27,753 ± 1550 | 103,500 ± 1705 | 5216 ± 354 | <0.0001 |
| Muc7 | 0.8 | 463 ± 78 | 400 ± 106 | 687 ± 163 | 1219 ± 740 | 494 ± 213 | 443 ± 143 | 370 ± 114 | 484 ± 115 | NA |
| Muc21 | 0.5 | 429 ± 113 | 559 ± 320 | 431 ± 5 | 1744 ± 315 | 1048 ± 303 | 598 ± 14 | 999 ± 208 | 450 ± 118 | NA |
| Mucl1 | 0.5 | 345 ± 96 | 1267 ± 780 | 613 ± 87 | 1032 ± 237 | 633 ± 83 | 549 ± 327 | 415 ± 116 | 511 ± 142 | NA |
| Muc4 | 0.4 | 1310 ± 339 | 470 ± 79 | 730 ± 372 | 358 ± 79 | 314 ± 59 | 18,487 ± 1343 | 496 ± 279 | 357 ± 110 | NA |
| Muc15 | 0.1 | 374 ± 130 | 929 ± 152 | 820 ± 224 | 540 ± 125 | 598 ± 121 | 4075 ± 536 | 19709 ± 895 | 611 ± 156 | NA |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woodfint, R.M.; Chen, P.R.; Ahn, J.; Suh, Y.; Hwang, S.; Lee, S.S.; Lee, K. Identification of the MUC2 Promoter as a Strong Promoter for Intestinal Gene Expression through Generation of Transgenic Quail Expressing GFP in Gut Epithelial Cells. Int. J. Mol. Sci. 2017, 18, 196. https://doi.org/10.3390/ijms18010196
Woodfint RM, Chen PR, Ahn J, Suh Y, Hwang S, Lee SS, Lee K. Identification of the MUC2 Promoter as a Strong Promoter for Intestinal Gene Expression through Generation of Transgenic Quail Expressing GFP in Gut Epithelial Cells. International Journal of Molecular Sciences. 2017; 18(1):196. https://doi.org/10.3390/ijms18010196
Chicago/Turabian StyleWoodfint, Rachel M., Paula R. Chen, Jinsoo Ahn, Yeunsu Suh, Seongsoo Hwang, Sang Suk Lee, and Kichoon Lee. 2017. "Identification of the MUC2 Promoter as a Strong Promoter for Intestinal Gene Expression through Generation of Transgenic Quail Expressing GFP in Gut Epithelial Cells" International Journal of Molecular Sciences 18, no. 1: 196. https://doi.org/10.3390/ijms18010196







