Highly Accurate Prediction of Protein-Protein Interactions via Incorporating Evolutionary Information and Physicochemical Characteristics
Abstract
:1. Introduction
2. Results and Discussion
2.1. Performance of the Proposed Method on Yeast and Helicobacter pylori (H. pylori) Datasets
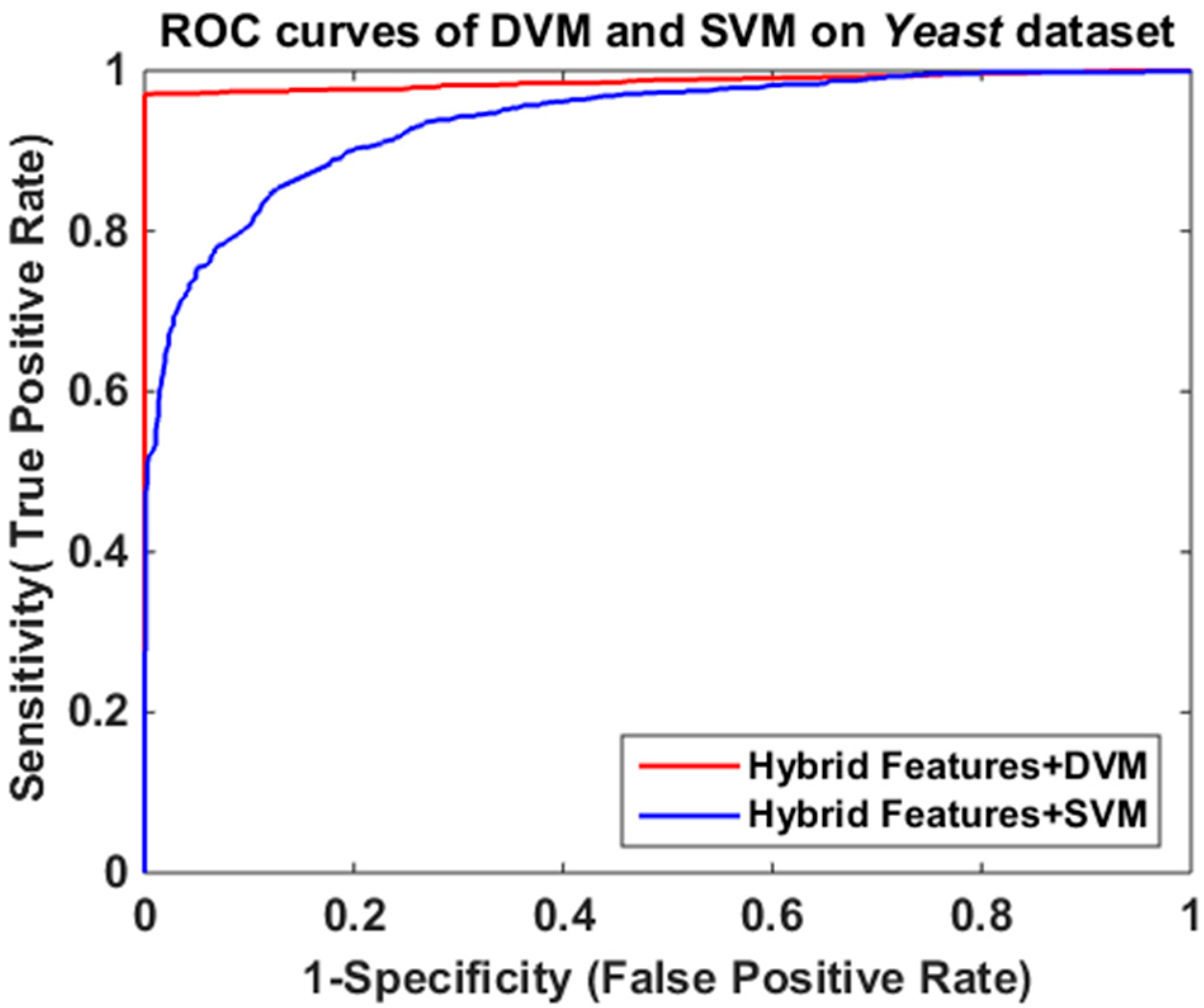
2.2. Comparison with SVM-Based Method
2.3. Comparison with Other Methods
3. Materials and Methods
3.1. Dataset
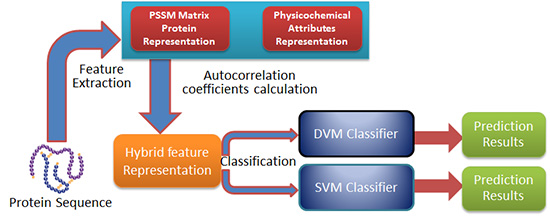
3.2. Feature Extraction
3.3. Discriminative Vector Machine
3.4. Procedure of the Proposed Method
3.5. Performance Evaluation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Huang, Y.A.; You, Z.H.; Chen, X.; Chan, K.; Luo, X. Sequence-based prediction of protein-protein interactions using weighted sparse representation model combined with global encoding. BMC Bioinform. 2016, 17, 184. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, L.; Wen, Z.; Li, M. Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences. Nucleic Acids Res. 2008, 36, 3025–3030. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; You, Z.-H.; Huang, D.-S. Increasing the reliability of protein-protein interaction networks via non-convex semantic embedding. Neurocomputing 2013, 121, 99–107. [Google Scholar] [CrossRef]
- Jia, J.; Liu, Z.; Xiao, X.; Liu, B.; Chou, K.C. iPPI-Esml: An ensemble classifier for identifying the interactions of proteins by incorporating their physicochemical properties and wavelet transforms into PseAAC. J. Theor. Biol. 2015, 377, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Chmielnicki, W.; Sta, K. A hybrid discriminative/generative approach to protein fold recognition. Neurocomputing 2012, 75, 194–198. [Google Scholar] [CrossRef]
- Ito, T.; Chiba, T.; Ozawa, R.; Yoshida, M.; Hattori, M.; Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 2001, 98, 4569–4574. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.; Bösche, M.; Krause, R.; Grandi, P.; Marzioch, M. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002, 415, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Bilgin, M.; Bangham, R.; Hall, D.; Casamayor, A. Global analysis of protein activities using proteome chips. Science 2001, 293, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Gruhler, A.; Heilbut, A.; Bader, G.; Moore, L. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 2002, 415, 180–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, R.; Fieldhouse, R.; Melo, A.; Correia, J.D.; Cordeiro, M.N.; Gumus, Z.H.; Costa, J.; Bonvin, A.M.; Moreira, I.S. A Machine Learning Approach for Hot-Spot Detection at Protein-Protein Interfaces. Int. J. Mol. Sci. 2016, 17, 1215. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhu, Y.; Liu, W. Combining Quantum-Behaved PSO and K2 Algorithm for Enhancing Gene Network Construction. Curr. Bioinform. 2013, 8, 133–137. [Google Scholar] [CrossRef]
- You, Z.-H.; Zhou, M.; Luo, X.; Li, S. Highly Efficient Framework for Predicting Interactions Between Proteins. IEEE Trans. Cybern. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S. Protein Sub-Nuclear Localization Based on Effective Fusion Representations and Dimension Reduction Algorithm LDA. Int. J. Mol. Sci. 2015, 16, 30343–30361. [Google Scholar] [CrossRef]
- Zheng, S.; Liu, W. An experimental comparison of gene selection by Lasso and Dantzig selector for cancer classification. Comput. Biol. Med. 2011, 41, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting protein-protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Z.; Gao, Y.; Zheng, Y.Y. Prediction of Protein-Protein Interactions Using Local Description of Amino Acid Sequence; Springer: Berlin, Germany, 2011; Volume 202, pp. 254–262. [Google Scholar]
- Zahiri, J.; Yaghoubi, O.; Mohammad-Noori, M.; Ebrahimpour, R.; Masoudi-Nejad, A. PPIevo: Protein-protein interaction prediction from PSSM based evolutionary information. Genomics 2013, 102, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Roe, D.; Faulon, J.L. Predicting protein-protein interactions using signature products. Bioinformatics 2005, 21, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; You, Z.-H.; Guo, H.; Luo, X.; Zhao, Z.-Q. Inverse-free Extreme Learning Machine with Optimal Information Updating. IEEE Trans. Cybern. 2016, 46, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Ong, Y.-S.; Zurada, J.M. Identification of Full and Partial Class Relevant Genes. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010, 7, 263–277. [Google Scholar] [PubMed]
- An, J.Y.; Meng, F.R.; You, Z.H.; Fang, Y.H.; Zhao, Y.J.; Zhang, M. Using the Relevance Vector Machine Model Combined with Local Phase Quantization to Predict Protein-Protein Interactions from Protein Sequences. BioMed Res. Int. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- An, J.Y.; You, Z.H.; Meng, F.R.; Xu, S.J.; Wang, Y. RVMAB: Using the Relevance Vector Machine Model Combined with Average Blocks to Predict the Interactions of Proteins from Protein Sequences. Int. J. Mol. Sci. 2016, 17, 757. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Ming, Z.; You, Z.; Li, S.; Xia, Y.; Leung, H. Improving network topology-based protein interactome mapping via collaborative filtering. Knowl. Based Syst. 2015, 90, 23–32. [Google Scholar] [CrossRef]
- Huang, Y.-A.; You, Z.-H.; Gao, X.; Wong, L.; Wang, L. Using Weighted Sparse Representation Model Combined with Discrete Cosine Transformation to Predict Protein-Protein Interactions from Protein Sequence. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Le, Y.; Zh, L.; Xi, J.; Wang, B. Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. BMC Bioinform. 2013, 14, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; You, Z.H.; Ming, Z.; Li, J.; Chen, X.; Huang, Y.A. Detection of Interactions between Proteins through Rotation Forest and Local Phase Quantization Descriptors. Int. J. Mol. Sci. 2016, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Liu, T.; Tao, D.; Sun, Z.; Tan, T. Representative Vector Machines: A unified framework for classical classifiers. IEEE Trans. Cybern. 2015, 46. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-Y.; Min, H.; Gui, J.; Zhu, L.; Lei, Y.-K. Face recognition via Weighted Sparse Representation. J. Vis. Commun. Image Represent. 2013, 24, 111–116. [Google Scholar] [CrossRef]
- LIBSVM—A Library for Support Vector Machines. Available online: http://www.csie.ntu.edu.tw/~cjlin/libsvm/ (accessed on 26 May 2016).
- Yang, L.; Xia, J.F.; Gui, J. Prediction of Protein-Protein Interactions from Protein Sequence Using Local Descriptors. Protein Pept. Lett. 2010, 17, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L. Fusion of classifiers for predicting protein–protein interactions. Neurocomputing 2005, 68, 289–296. [Google Scholar] [CrossRef]
- Nanni, L. Hyperplanes for predicting protein-protein interactions. Neurocomputing 2005, 69, 257–263. [Google Scholar] [CrossRef]
- Nanni, L.; Lumini, A. An ensemble of K-local hyperplanes for predicting protein-protein interactions. Bioinformatics 2006, 22, 1207–1210. [Google Scholar] [CrossRef] [PubMed]
- Xenarios, I.; Salwínski, L.; Duan, X.; Higney, P.; Kim, S. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002, 30, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jaroszewski, L.; Godzik, A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 2001, 17, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, Y.H.; Gromiha, M.M. Application of amino acid occurrence for discriminating different folding types of globular proteins. BMC Bioinform. 2007, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- AAindex. Available online: http://www.genome.jp/aaindex/ (accessed on 20 May 2016).
- Kawashima, S.; Ogata, H.; Kanehisa, M. AAindex: Amino Acid Index Database. Nucleic Acids Res. 1999, 27, 368–369. [Google Scholar]
- Raicar, G.; Saini, H.; Dehzangi, A.; Lal, S.; Sharma, A. Improving protein fold recognition and structural class prediction accuracies using physicochemical properties of amino acids. J. Theor. Biol. 2016, 402, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct. Funct. Bioinform. 2001, 43, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, D.N.; Karakasidis, T.E.; Megaritis, A.C. A short survey on genetic sequences, chou’s pseudo amino acid composition and its combination with fuzzy set theory. Open Bioinform. J. 2013, 7, 41–48. [Google Scholar] [CrossRef]
- Altschul, S.; Koonin, E. Iterated profile searches with PSI-BLAST—A tool for discovery in protein databases. Trends Biochem. Sci. 1998, 23, 444–447. [Google Scholar] [CrossRef]
- Liu, W.; Pokharel, P.P.; Principe, J.C. Correntropy: Properties and Applications in Non-Gaussian Signal Processing. IEEE Trans. Signal. Process. 2007, 55, 5286–5298. [Google Scholar] [CrossRef]
- Olive, D.J. A resistant estimator of multivariate location and dispersion. Comput. Stat. Data Anal. 2004, 46, 93–102. [Google Scholar] [CrossRef]
- He, R.; Zheng, W.; Hu, B. Maximum Correntropy Criterion for Robust Face Recognition. IEEE Trans. Softw. Eng. 2011, 33, 1561–1576. [Google Scholar]

| Testing Set | Acc (%) | Sen (%) | Pre (%) | MCC (%) |
|---|---|---|---|---|
| 1 | 93.52 | 93.07 | 98.34 | 88.94 |
| 2 | 94.76 | 92.41 | 96.56 | 87.57 |
| 3 | 93.83 | 93.64 | 95.68 | 87.61 |
| 4 | 94.43 | 93.52 | 96.67 | 90.02 |
| 5 | 95.21 | 92.19 | 95.33 | 91.19 |
| Average | 94.35 ± 0.68 | 92.97 ± 0.65 | 96.52 ± 1.17 | 89.07 ± 1.56 |
| Testing Set | Acc (%) | Sen (%) | Pre (%) | MCC (%) |
|---|---|---|---|---|
| 1 | 92.81 | 92.28 | 93.40 | 83.32 |
| 2 | 89.59 | 90.73 | 91.73 | 81.08 |
| 3 | 90.82 | 93.37 | 90.19 | 84.51 |
| 4 | 91.06 | 89.64 | 89.27 | 83.61 |
| 5 | 88.75 | 90.59 | 89.12 | 81.43 |
| Average | 90.61 ± 1.55 | 91.32 ± 1.48 | 90.74 ± 1.81 | 82.79 ± 1.47 |
| Model | Testing Set | Acc (%) | Sen (%) | Pre (%) | MCC (%) |
|---|---|---|---|---|---|
| SVM | 1 | 85.12 | 84.87 | 86.34 | 75.92 |
| 2 | 86.16 | 83.91 | 85.36 | 74.99 | |
| 3 | 87.96 | 85.64 | 86.61 | 77.58 | |
| 4 | 85.42 | 85.80 | 88.67 | 75.02 | |
| 5 | 84.21 | 86.70 | 85.33 | 74.76 | |
| Average | 85.77 ± 1.41 | 85.38 ± 1.05 | 86.46 ± 1.36 | 75.65 ± 1.16 | |
| DVM | 1 | 93.52 | 93.07 | 98.34 | 88.94 |
| 2 | 94.76 | 92.41 | 96.56 | 87.57 | |
| 3 | 93.83 | 93.64 | 95.68 | 87.61 | |
| 4 | 94.43 | 93.52 | 96.67 | 90.02 | |
| 5 | 95.21 | 92.19 | 95.33 | 91.19 | |
| Average | 94.35 ± 0.68 | 92.97 ± 0.65 | 96.52 ± 1.17 | 89.07 ± 1.56 |
| Model | Testing Set | Acc (%) | Sen (%) | Pre (%) | MCC (%) |
|---|---|---|---|---|---|
| Guo [2] | ACC | 89.33 ± 2.67 | 89.93 ± 3.68 | 88.87 ± 6.16 | N/A |
| AC | 87.36 ± 1.38 | 87.30 ± 4.68 | 87.82 ± 4.33 | N/A | |
| Yang [30] | Cod1 | 75.08 ± 1.13 | 75.81 ± 1.20 | 74.75 ± 1.23 | N/A |
| Cod2 | 80.04 ± 1.06 | 76.77 ± 0.69 | 82.17 ± 1.35 | N/A | |
| Cod3 | 80.41 ± 0.47 | 78.14 ± 0.90 | 81.66 ± 0.99 | N/A | |
| Cod4 | 86.15 ± 1.17 | 81.03 ± 1.74 | 90.24 ± 1.34 | N/A | |
| You [25] | PCA-EELM | 87.00 ± 0.29 | 86.15 ± 0.43 | 87.59 ± 0.32 | 77.36 ± 0.44 |
| Wong [26] | RF + PR-LPQ | 93.92 ± 0.36 | 91.10 ± 0.31 | 96.45 ± 0.45 | 88.56 ± 0.63 |
| Proposed Method | DVM | 94.35 ± 0.67 | 92.97 ± 0.51 | 96.52 ± 0.57 | 89.07 ± 1.30 |
| Model | Acc (%) | Sen (%) | Pre (%) | MCC (%) |
|---|---|---|---|---|
| Nanni [31] | 83.00 | 86.00 | 85.10 | N/A |
| Nanni [32] | 84.00 | 86.00 | 84.00 | N/A |
| Nanni and Lumini [33] | 86.60 | 86.70 | 85.00 | N/A |
| You [25] | 87.50 | 88.95 | 86.15 | 78.13 |
| Martin [18] | 83.40 | 79.90 | 85.70 | N/A |
| Wong [26] | 89.47 | 89.18 | 89.63 | 81.00 |
| Proposed Method | 90.61 | 91.32 | 90.74 | 82.79 |
| Amino Acid Name | Hydrophobicity | Polarity | Polarizability | van der Waals Volume |
|---|---|---|---|---|
| Alanine | 0.61 | 8.1 | 0.046 | 1.00 |
| Arginine | 0.60 | 10.5 | 0.291 | 6.13 |
| Asparagine | 0.06 | 11.6 | 0.134 | 2.95 |
| Aspartic Acid | 0.46 | 13.0 | 0.105 | 2.78 |
| Cysteine | 1.07 | 5.5 | 0.128 | 2.43 |
| Glutamine | 0.0 | 10.5 | 0.180 | 3.95 |
| Glutamic Acid | 0.47 | 12.3 | 0.151 | 3.78 |
| Glycine | 0.07 | 9.0 | 0.000 | 0.00 |
| Histidine | 0.61 | 10.4 | 0.230 | 4.66 |
| Isoleucine | 2.22 | 5.2 | 0.186 | 4.00 |
| Leucine | 1.53 | 4.9 | 0.186 | 4.00 |
| Lysine | 1.15 | 11.3 | 0.219 | 4.77 |
| Methionine | 1.18 | 5.7 | 0.221 | 4.43 |
| Phenylalanine | 2.02 | 5.2 | 0.290 | 5.89 |
| Proline | 1.95 | 8.0 | 0.131 | 2.72 |
| Serine | 0.05 | 9.2 | 0.062 | 1.60 |
| Threonine | 0.05 | 8.6 | 0.108 | 2.60 |
| Tryptophan | 2.65 | 5.4 | 0.409 | 8.08 |
| Tyrosine | 1.88 | 6.2 | 0.298 | 6.47 |
| Valine | 1.32 | 5.9 | 0.140 | 3.00 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.-W.; You, Z.-H.; Chen, X.; Gui, J.; Nie, R. Highly Accurate Prediction of Protein-Protein Interactions via Incorporating Evolutionary Information and Physicochemical Characteristics. Int. J. Mol. Sci. 2016, 17, 1396. https://doi.org/10.3390/ijms17091396
Li Z-W, You Z-H, Chen X, Gui J, Nie R. Highly Accurate Prediction of Protein-Protein Interactions via Incorporating Evolutionary Information and Physicochemical Characteristics. International Journal of Molecular Sciences. 2016; 17(9):1396. https://doi.org/10.3390/ijms17091396
Chicago/Turabian StyleLi, Zheng-Wei, Zhu-Hong You, Xing Chen, Jie Gui, and Ru Nie. 2016. "Highly Accurate Prediction of Protein-Protein Interactions via Incorporating Evolutionary Information and Physicochemical Characteristics" International Journal of Molecular Sciences 17, no. 9: 1396. https://doi.org/10.3390/ijms17091396
APA StyleLi, Z.-W., You, Z.-H., Chen, X., Gui, J., & Nie, R. (2016). Highly Accurate Prediction of Protein-Protein Interactions via Incorporating Evolutionary Information and Physicochemical Characteristics. International Journal of Molecular Sciences, 17(9), 1396. https://doi.org/10.3390/ijms17091396