Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m6A Modifications in Adipose and Skeletal Muscle Tissues in Offspring
Abstract
:1. Introduction
2. Results
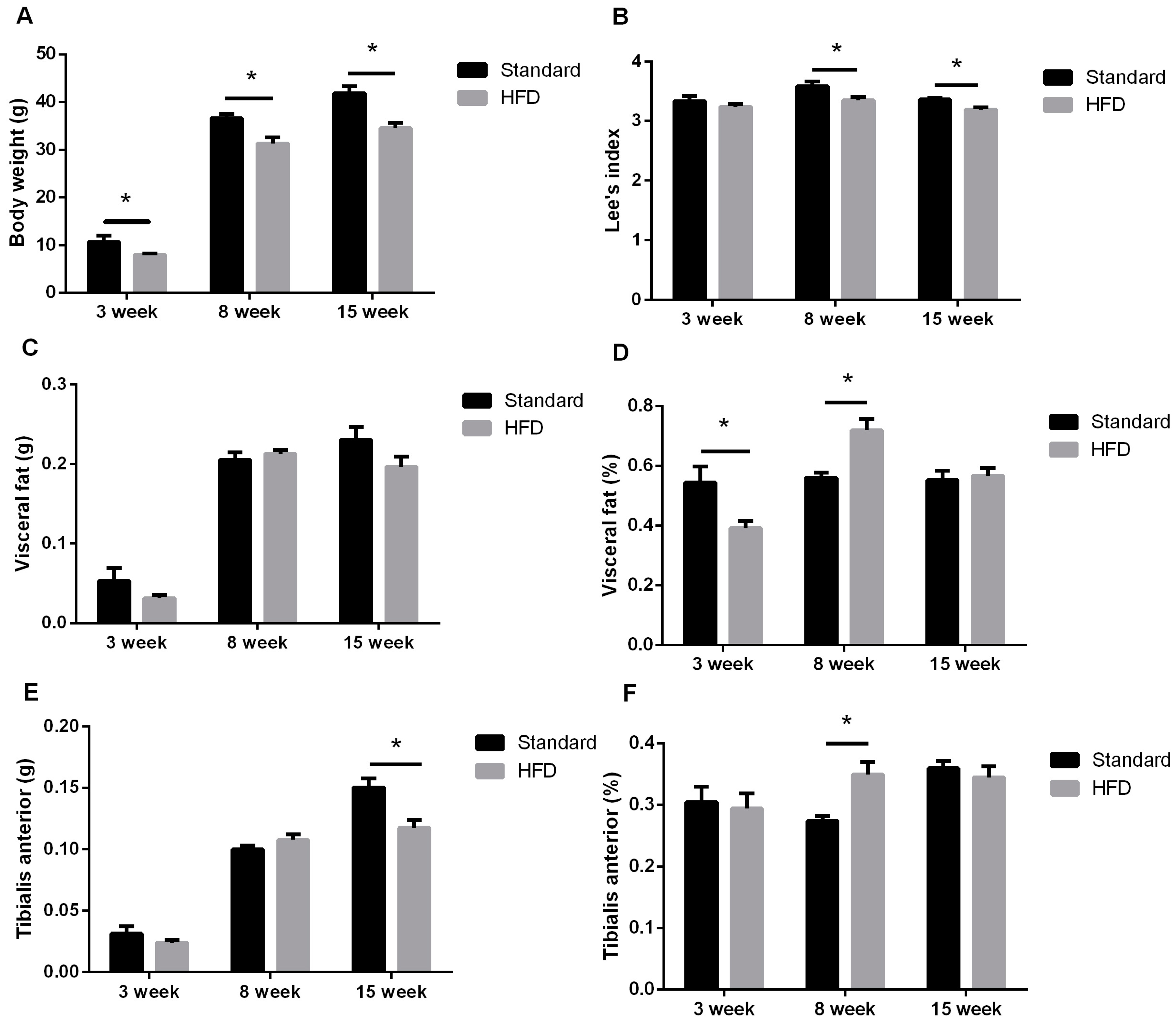
2.1. High-Fat Diet Caused Obesity and Glucose Intolerance in Dams
2.2. Maternal High-Fat Intake Impaired the Growth of Male Litters
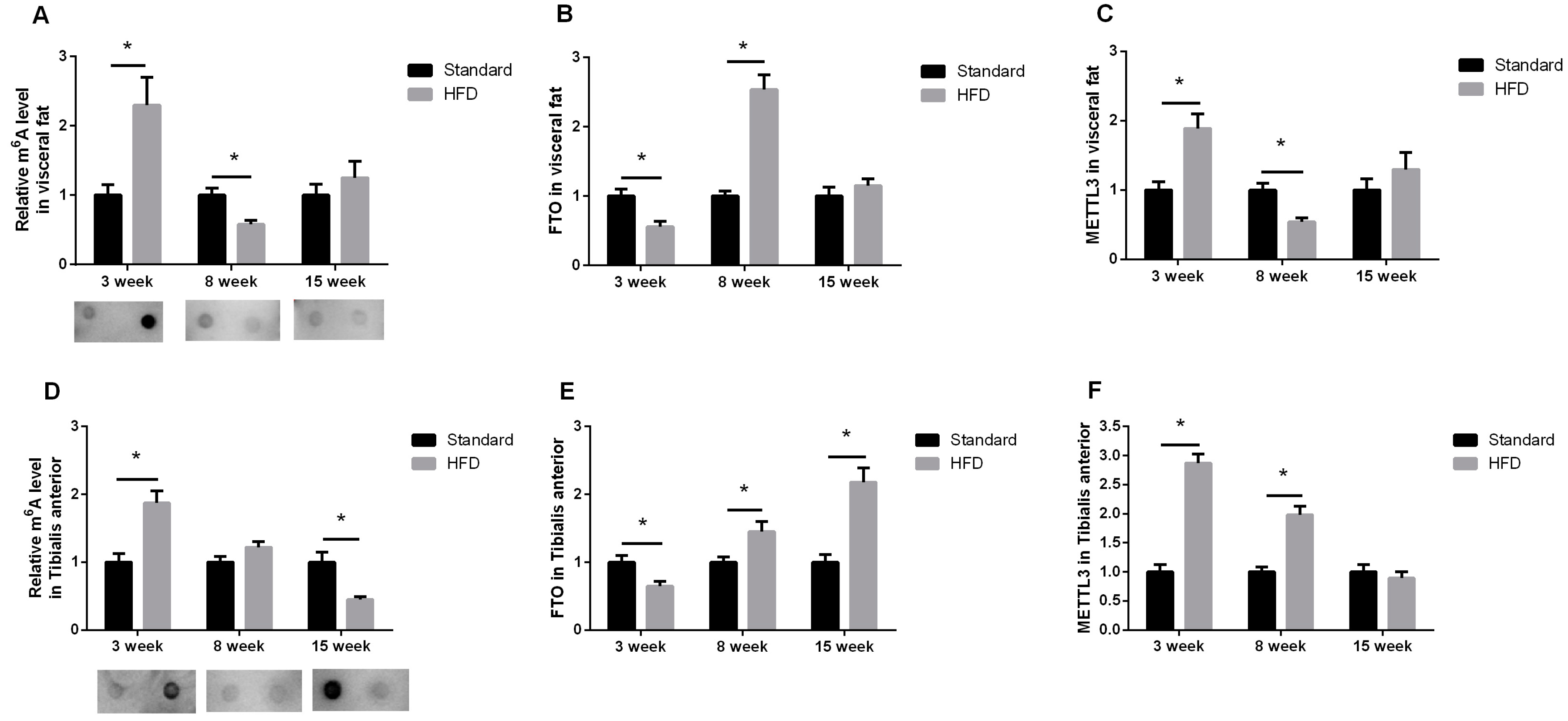
2.3. Maternal High-Fat Diet Altered the m6A Pattern in Fat and Skeletal Muscle in a Development-Dependent Way
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
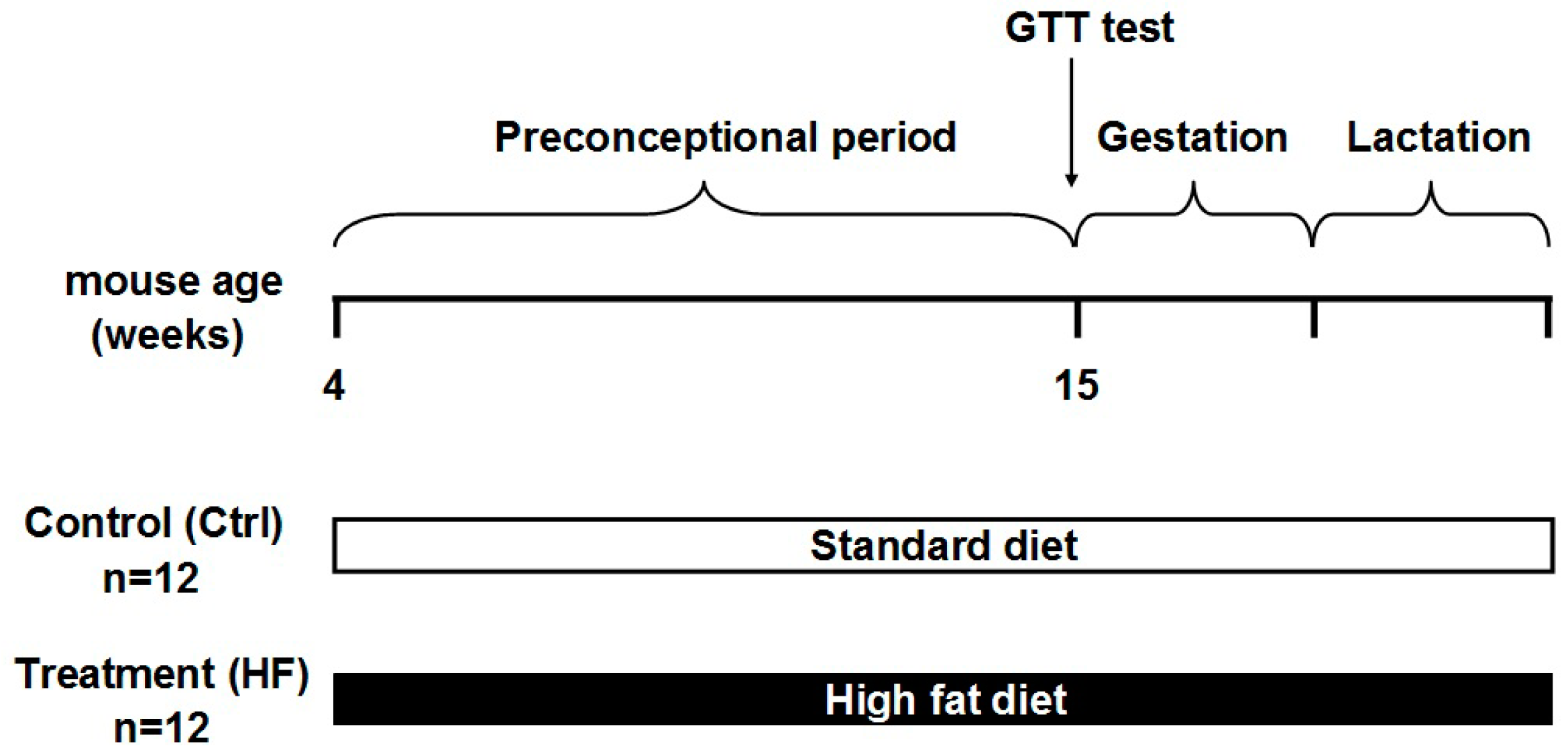
4.2. Animals and Diet
4.3. Glucose Tolerance Test
4.4. RNA Isolation
4.5. RNA Dot-Blot
4.6. qRT-PCR
4.7. Insulin Sensitity Assay
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yokomizo, H.; Inoguchi, T.; Sonoda, N.; Sakaki, Y.; Maeda, Y.; Inoue, T.; Hirata, E.; Takei, R.; Ikeda, N.; Fujii, M.; et al. Maternal high-fat diet induces insulin resistance and deterioration of pancreatic β-cell function in adult offspring with sex differences in mice. Am. J. Physiol. Endocrinol. Metab. 2014, 306, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cao, M.; Yang, M.; Lin, Y.; Che, L.; Fang, Z.; Xu, S.; Feng, B.; Li, J.; Wu, D. Intra-uterine undernutrition amplifies age-associated glucose intolerance in pigs via altered DNA methylation at muscle GLUT4 promoter. Br. J. Nutr. 2016, 116, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Marco, A.; Kisliouk, T.; Tabachnik, T.; Weller, A.; Meiri, N. DNA CpG methylation (5mC) and its derivative (5hmC) alter histone post translational modifications at the POMC promoter, affecting the impact of perinatal diet on leanness and obesity of the offspring. Diabetes 2016. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Li, H.-B.; Yin, Z.; Flavell, R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA m6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. AlkBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.; Girard, C.A.; Tung, Y.-C.L.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.H.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The obesity-associated FTO gene encodes a 2-oxoglutarate–dependent nucleic acid demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Aik, W.; Scotti, J.S.; Choi, H.; Gong, L.; Demetriades, M.; Schofield, C.J.; McDonough, M.A. Structure of human RNA m6-methyladenine demethylase AlkBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014, 42, 4741–4754. [Google Scholar] [CrossRef] [PubMed]
- Grunnet, L.G.; Nilsson, E.; Ling, C.; Hansen, T.; Pedersen, O.; Groop, L.; Vaag, A.; Poulsen, P. Regulation and function of FTO mRNA expression in human skeletal muscle and subcutaneous adipose tissue. Diabetes 2009, 58, 2402–2408. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.; Ross, M.G. Fetal programming of adipose tissue: Effects of IUGR and maternal obesity/high fat diet. Semin. Reprod. Med. 2011, 29, 237–245. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, R.E.K.; Castelli, L.M.; Miotto, P.M.; Frendo-Cumbo, S.; Milburn, A.; Roy, B.D.; LeBlanc, P.J.; Ward, W.E.; Peters, S.J. A maternal high fat diet has long-lasting effects on skeletal muscle lipid and PLIN protein content in rat offspring at young adulthood. Lipids 2015, 50, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Sasson, I.; Vitins, A.; Mainigi, M.; Moley, K.; Simmons, R. Pre-gestational vs gestational exposure to maternal obesity differentially programs the offspring in mice. Diabetologia 2015, 58, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Gray, C.; Vickers, M.H.; Segovia, S.A.; Zhang, X.D.; Reynolds, C.M. A maternal high fat diet programmes endothelial function and cardiovascular status in adult male offspring independent of body weight, which is reversed by maternal conjugated linoleic acid (CLA) supplementation. PLoS ONE 2015, 10, e0115994. [Google Scholar]
- Desai, M.; Jellyman, J.K.; Han, G.; Beall, M.; Lane, R.H.; Ross, M.G. Rat maternal obesity and high fat diet program offspring metabolic syndrome. Am. J. Obstet. Gynecol. 2014, 211, 237.e213–237.e231. [Google Scholar] [CrossRef] [PubMed]
- Latouche, C.; Heywood, S.E.; Henry, S.L.; Ziemann, M.; Lazarus, R.; El-Osta, A.; Armitage, J.A.; Kingwell, B.A. Maternal overnutrition programs changes in the expression of skeletal muscle genes that are associated with insulin resistance and defects of oxidative phosphorylation in adult male rat offspring. J. Nutr. 2014, 144, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Masuyama, H.; Mitsui, T.; Eguchi, T.; Tamada, S.; Hiramatsu, Y. The effects of paternal high-fat diet exposure on offspring metabolism with epigenetic changes in the mouse adiponectin and leptin gene promoter. Am. J. Physiol. Endocrinol. Metab. 2016, 311, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mrna methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, L.; Chen, J.; Wang, Y. mRNA m6A methylation downregulates adipogenesis in porcine adipocytes. Biochem. Biophys. Res. Commun. 2015, 459, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.; Ma, J.; Guo, F.; Cao, Q.; Zhang, Y.; Zhou, B.; Chai, J.; Zhao, W.; Zhao, R. The demethylase activity of FTO (fat mass and obesity associated protein) is required for preadipocyte differentiation. PLoS ONE 2015, 10, e0133788. [Google Scholar] [CrossRef]
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Bruning, J.C.; Nolan, P.M.; Ashcroft, F.M.; et al. Overexpression of FTO leads to increased food intake and results in obesity. Nat. Genet. 2010, 42, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Bruning, J.C.; Ruther, U.; Horsthemke, B. Inactivation of the Fto gene protects from obesity. Nature 2009, 458, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Berulava, T.; Ziehe, M.; Klein-Hitpass, L.; Mladenov, E.; Thomale, J.; Rüther, U.; Horsthemke, B. FTO levels affect RNA modification and the transcriptome. Eur. J. Hum. Genet. 2013, 21, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Merkestein, M.; McTaggart, J.S.; Lee, S.; Kramer, H.B.; McMurray, F.; Lafond, M.; Boutens, L.; Cox, R.; Ashcroft, F.M. Changes in gene expression associated with Fto overexpression in mice. PLoS ONE 2014, 9, e97162. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Song, W.T.; Shu, J.T.; Tao, Z.Y.; Zhu, W.Q.; Di, C.; Li, H.F. Tissue- and breed-specific expression of the chicken fat mass- and obesity-associated gene (FTO). Genet. Mol. Res. 2015, 14, 10500–10506. [Google Scholar] [CrossRef] [PubMed]
- Bravard, A.; Lefai, E.; Meugnier, E.; Pesenti, S.; Disse, E.; Vouillarmet, J.; Peretti, N.; Rabasa-Lhoret, R.; Laville, M.; Vidal, H.; et al. FTO is increased in muscle during type 2 diabetes, and its overexpression in myotubes alters insulin signaling, enhances lipogenesis and ros production, and induces mitochondrial dysfunction. Diabetes 2011, 60, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Cunha, F.D.S.; Dalle Molle, R.; Portella, A.K.; Benetti, C.D.S.; Noschang, C.; Goldani, M.Z.; Silveira, P.P. Both food restriction and high-fat diet during gestation induce low birth weight and altered physical activity in adult rat offspring: The “similarities in the inequalities” model. PLoS ONE 2015, 10, e0118586. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Wang, H.; Cui, H.; Chen, H.; Pan, Y.-X. Early-life exposure to high-fat diet may predispose rats to gender-specific hepatic fat accumulation by programming PEPCK expression. J. Nutr. Biochem. 2014, 26, 433–440. [Google Scholar] [CrossRef] [PubMed]

| Groups | FBG (mmol/L) | Fins (mIU/L) | HOMA-IR |
|---|---|---|---|
| Standard group | 5.18 ± 0.25 | 9.76 ± 0.87 | 2.20 ± 0.15 |
| HFD group | 5.67 ± 0.26 | 10.12 ± 0.68 | 2.51 ± 0.16 |
| Genes | Access No. | Sequences (5′→3′) | Amplicon Size |
|---|---|---|---|
| FTO | NM_011936 | F: CCGTCCTGCGATGATGAAGT | 119 bp |
| R: CCCATGCCGAAATAGGGCTC | |||
| METTLS | NM_019721 | F: GAGTTGATTGAGGTAAAGCGAGG | 75 bp |
| R: GGAGTGGTCAGCGTAAGTTACA | |||
| β-actin | NM_007393 | F: GGCTGTATTCCCCTCCATCG | 154 bp |
| R: CCAGTTGGTAACAATGCCATGT |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yang, J.; Zhu, Y.; Liu, Y.; Shi, X.; Yang, G. Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m6A Modifications in Adipose and Skeletal Muscle Tissues in Offspring. Int. J. Mol. Sci. 2016, 17, 1336. https://doi.org/10.3390/ijms17081336
Li X, Yang J, Zhu Y, Liu Y, Shi X, Yang G. Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m6A Modifications in Adipose and Skeletal Muscle Tissues in Offspring. International Journal of Molecular Sciences. 2016; 17(8):1336. https://doi.org/10.3390/ijms17081336
Chicago/Turabian StyleLi, Xiao, Jing Yang, Youbo Zhu, Yuan Liu, Xin’e Shi, and Gongshe Yang. 2016. "Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m6A Modifications in Adipose and Skeletal Muscle Tissues in Offspring" International Journal of Molecular Sciences 17, no. 8: 1336. https://doi.org/10.3390/ijms17081336
APA StyleLi, X., Yang, J., Zhu, Y., Liu, Y., Shi, X., & Yang, G. (2016). Mouse Maternal High-Fat Intake Dynamically Programmed mRNA m6A Modifications in Adipose and Skeletal Muscle Tissues in Offspring. International Journal of Molecular Sciences, 17(8), 1336. https://doi.org/10.3390/ijms17081336






