Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications
Abstract
:1. Introduction
2. Deep Artificial Neural Networks in Pharmacology and Bioinformatics
2.1. Deep Auto-Encoder Networks
2.1.1. Pharmacology
2.1.2. Bioinformatics
2.2. Deep Convolutional Neural Networks
2.2.1. Pharmacology
2.2.2. Bioinformatics
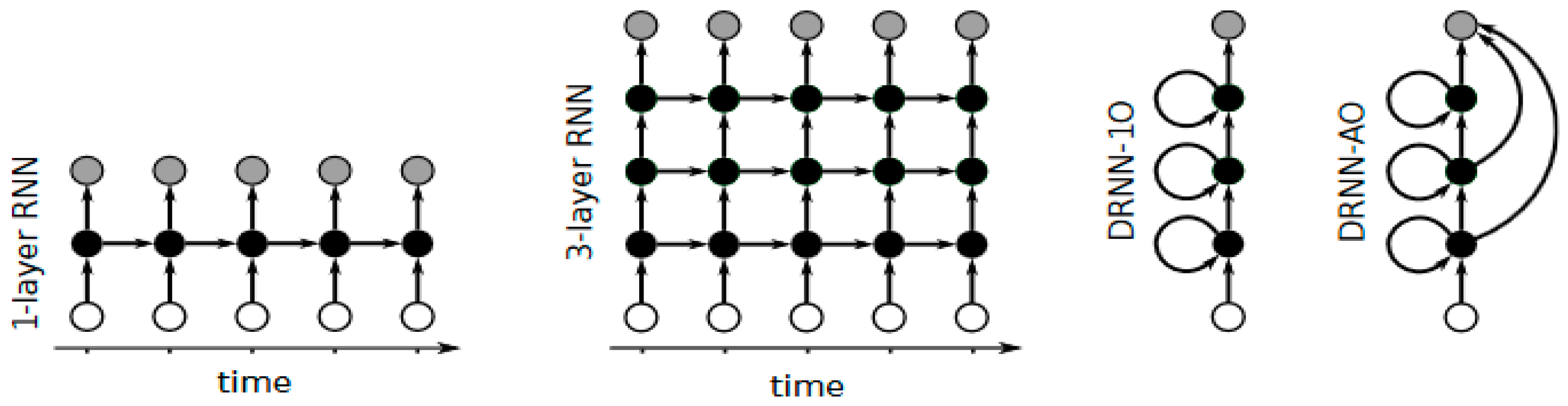
2.3. Deep Recurrent Neural Networks
2.3.1. Pharmacology
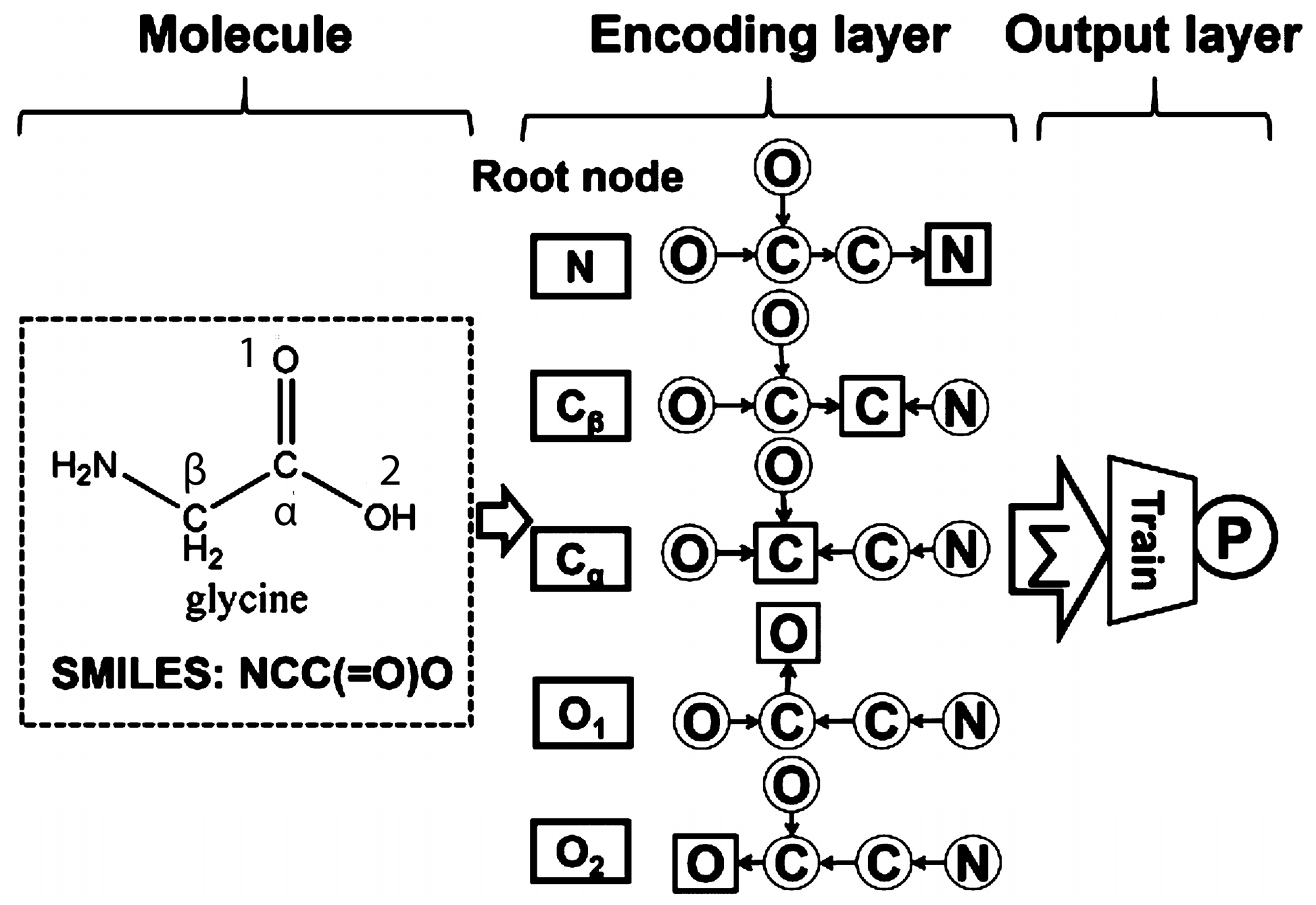
2.3.2. Bioinformatics
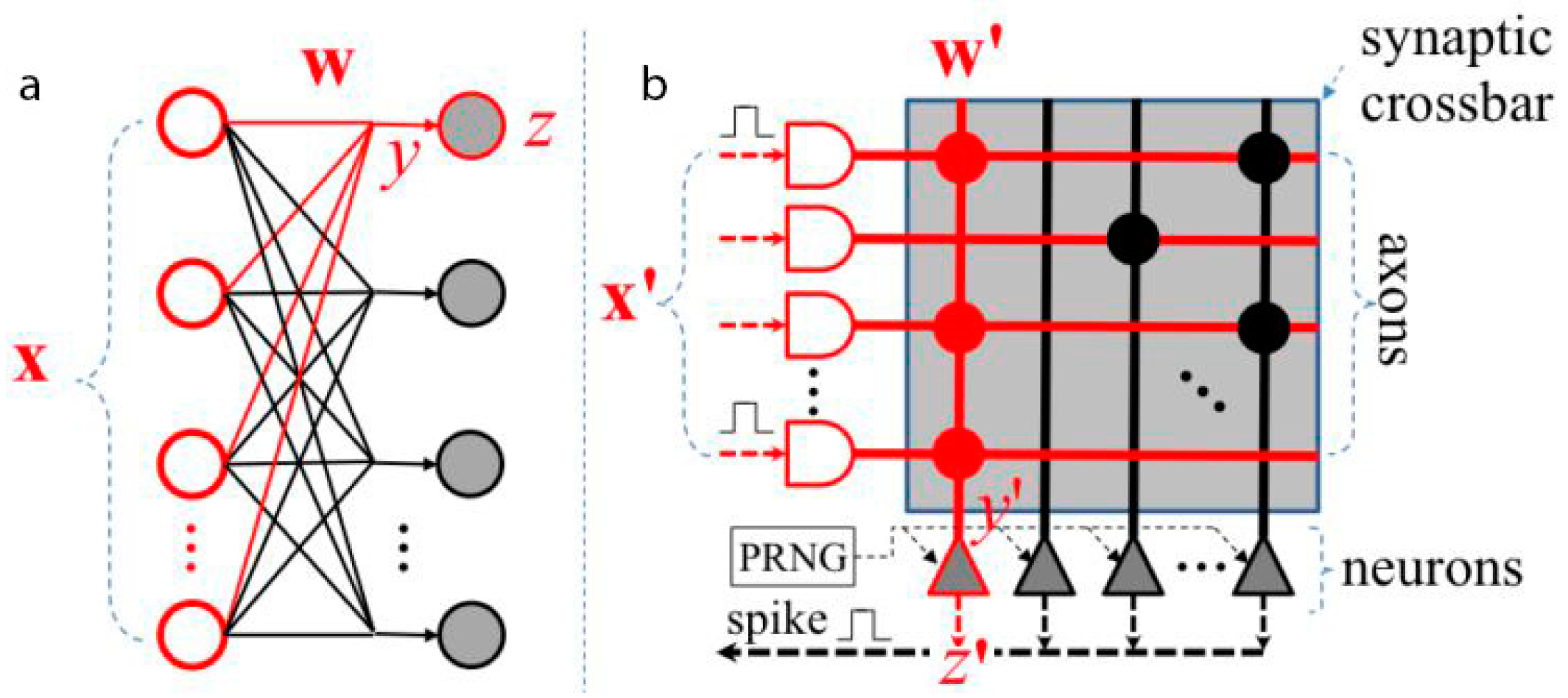
3. Neuromorphic Chips
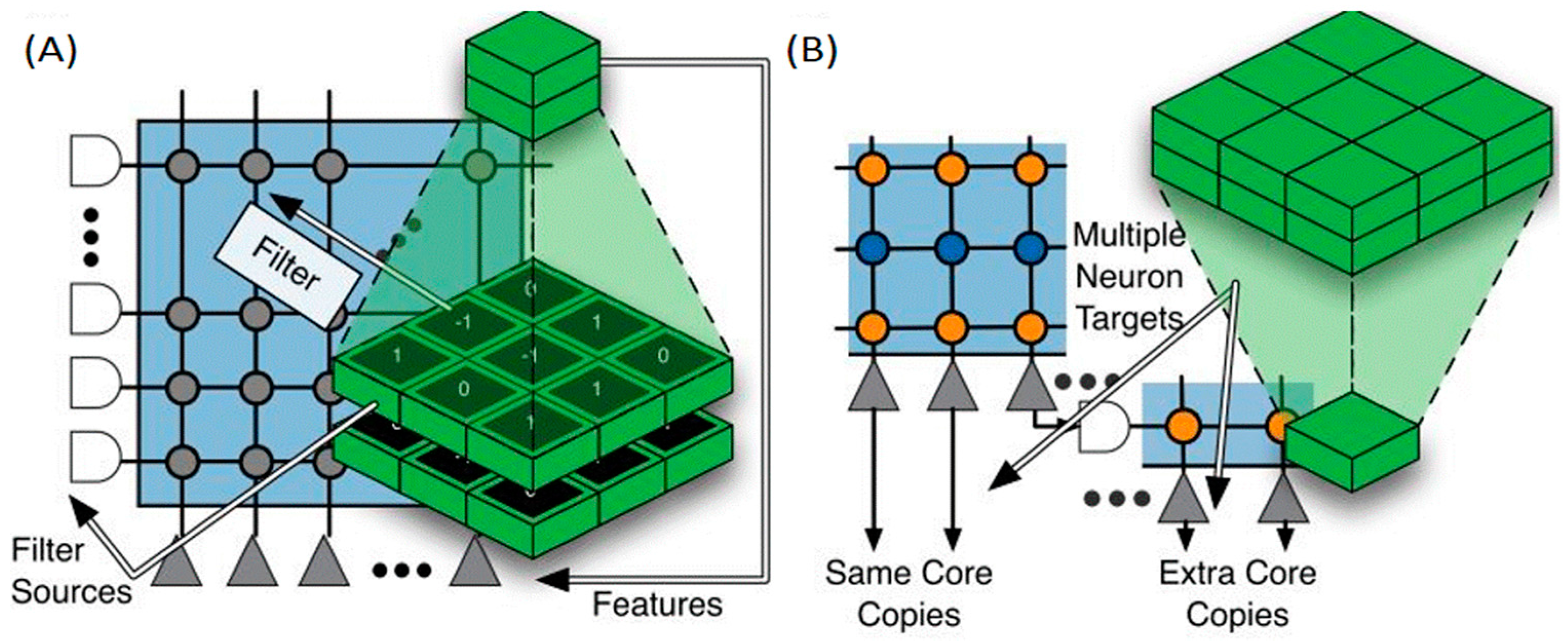
3.1. TrueNorth International Business Machines (IBM)
3.2. SpiNNaker. University of Manchester
- Deep Belief Networks: These networks of deep learning may be implemented, obtaining an accuracy rate of 95% in the classification of the MNIST database of handwritten digits. Results of 0.06% less accuracy than with the software implementation are obtained, whereas the consumption is only 0.3 W [36,110].
- Convolutional Neural Networks: This type of networks has the characteristic of sharing the same value of weights for many neuron-to-neuron connections, which reduces the amount of memory required to store the synaptic weights. A five-layer deep learning network is implemented to recognize symbols which are obtained through a Dynamic Vision Sensor. Each ARM core can accommodate 2048 neurons. The full chip could contain up to 32,000 neurons. A particular ConvNet architecture was implemented in SpiNNaker for visual object recognition, like poker card symbol classification [111].
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ADME | Absorption, Distribution, Metabolism, and Excretion |
| AER | Address Event Representation |
| ANGN | Artificial Neuron-Glia Networks |
| ANN | Artificial Neural Networks |
| AUC | Area Under the Receiver Operating Characteristic Curve |
| CASP | Critical Assessment of protein Structure |
| CNN | Convolutional Neural Networks |
| CPU | Central Processing Unit |
| CUDA | Compute Unified Device Architecture |
| DAEN | Deep Auto-Encoder Networks |
| DANAN | Deep Artificial Neuron–Astrocyte Networks |
| DBN | Deep Belief Networks |
| DCNN | Deep Convolution Neural Networks |
| DFNN | Deep Feedforward Neural Networks |
| DL | Deep Learning |
| DNN | Deep Artificial Neural Networks |
| DBM | Deep Boltzmann Machines |
| DRNN | Deep Recurrent Neural Networks |
| ECFP4 | Extended Connectivity Fingerprints |
| GPGPUs | General-Purpose Graphical Processing Units |
| GPU | Graphical Processing Unit |
| ML | Machine Learning |
| QSAR | Quantitative Structure–Activity Relationship |
| QSPkR | Quantitative Structure–Pharmacokinetic Relationship |
| QSPR | Quantitative Structure–Property Relationships |
| QSTR | Quantitative Structure–Toxicity Relationship |
| SANN | Spiking Artificial Neural Network |
| SVM | Support Vector Machines |
| VLSI | Very Large Scale Integration |
| VS | Virtual Screening |
References
- Gawehn, E.; Hiss, J.A.; Schneider, G. Deep learning in drug discovery. Mol. Inform. 2016, 35, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, M.; Suchacz, B. Artificial neural networks: Theoretical background and pharmaceutical applications: A review. J. AOAC Int. 2012, 95, 652–668. [Google Scholar] [CrossRef] [PubMed]
- Gertrudes, J.C.; Maltarollo, V.G.; Silva, R.A.; Oliveira, P.R.; Honório, K.M.; da Silva, A.B.F. Machine learning techniques and drug design. Curr. Med. Chem. 2012, 19, 4289–4297. [Google Scholar] [CrossRef] [PubMed]
- Puri, M.; Pathak, Y.; Sutariya, V.K.; Tipparaju, S.; Moreno, W. Artificial Neural Network for Drug Design, Delivery and Disposition; Elsevier Science: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Yee, L.C.; Wei, Y.C. Current modeling methods used in QSAR/QSPR. In Statistical Modelling of Molecular Descriptors in QSAR/QSPR; John Wiley & Sons: Hoboken, NJ, USA, 2012; Volume 10, pp. 1–31. [Google Scholar]
- Qian, N.; Sejnowski, T.J. Predicting the secondary structure of globular proteins using neural network models. J. Mol. Biol. 1988, 202, 865–884. [Google Scholar] [CrossRef]
- Aoyama, T.; Suzuki, Y.; Ichikawa, H. Neural networks applied to structure-activity relationships. J. Med. Chem. 1990, 33, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Wikel, J.H.; Dow, E.R. The use of neural networks for variable selection in QSAR. Bioorg. Med. Chem. Lett. 1993, 3, 645–651. [Google Scholar] [CrossRef]
- Tetko, I.V.; Tanchuk, V.Y.; Chentsova, N.P.; Antonenko, S.V.; Poda, G.I.; Kukhar, V.P.; Luik, A.I. HIV-1 reverse transcriptase inhibitor design using artificial neural networks. J. Med. Chem. 1994, 37, 2520–2526. [Google Scholar] [CrossRef] [PubMed]
- Kovalishyn, V.V.; Tetko, I.V.; Luik, A.I.; Kholodovych, V.V.; Villa, A.E.P.; Livingstone, D.J. Neural network studies. 3. variable selection in the cascade-correlation learning architecture. J. Chem. Inf. Comput. Sci. 1998, 38, 651–659. [Google Scholar] [CrossRef]
- Yousefinejad, S.; Hemmateenejad, B. Chemometrics tools in QSAR/QSPR studies: A historical perspective. Chemom. Intell. Lab. Syst. 2015, 149, 177–204. [Google Scholar] [CrossRef]
- Lavecchia, A. Machine-learning approaches in drug discovery: Methods and applications. Drug Discov. Today 2015, 20, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Vidyasagar, M. Identifying predictive features in drug response using machine learning: Opportunities and challenges. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Dobchev, D.A.; Pillai, G.G.; Karelson, M. In silico machine learning methods in drug development. Curr. Top. Med. Chem. 2014, 14, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Omer, A.; Singh, P.; Yadav, N.K.; Singh, R.K. An overview of data mining algorithms in drug induced toxicity prediction. Mini Rev. Med. Chem. 2014, 14, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Pandini, A.; Fraccalvieri, D.; Bonati, L. Artificial neural networks for efficient clustering of conformational ensembles and their potential for medicinal chemistry. Curr. Top. Med. Chem. 2013, 13, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, K.; Lyons, J.; Heffernan, R. A short review of deep learning neural networks in protein structure prediction problems. Adv. Tech. Biol. Med. 2015. [Google Scholar] [CrossRef]
- Cheng, F. Applications of artificial neural network modeling in drug discovery. Clin. Exp. Pharmacol. 2012. [Google Scholar] [CrossRef]
- Udemy Blog. Available online: https://blog.udemy.com/wp-content/uploads/2014/04/Hadoop-Ecosystem.jpg (accessed on 13 May 2016).
- Neural Networks and Deep Learning. Available online: http://neuralnetworksanddeeplearning.com/chap5.html (accessed on 13 May 2016).
- Unsupervised Feature Learning and Deep Learning. Available online: http://ufldl.stanford.edu/wiki/index.php/Deep_Networks: Overview#Diffusion_of_gradients (accessed on 13 May 2016).
- Furber, S.B. Brain-Inspired Computing. IET Comput. Dig. Tech. 2016. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Deng, L. A tutorial survey of architectures, algorithms, and applications for deep learning. APSIPA Trans. Signal Inf. Process. 2014, 3, e2. [Google Scholar] [CrossRef]
- Deng, L. Deep learning: methods and applications. Found. Trends Signal Process. 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Wang, H.; Raj, B. A Survey: Time Travel in Deep Learning Space: An Introduction to DEEP Learning Models and How Deep Learning Models Evolved from the Initial Ideas. Available online: http://arxiv.org/abs/1510.04781 (accessed on 13 May 2016).
- Lipton, Z.C. A Critical Review of Recurrent Neural Networks for Sequence Learning. Available online: http://arXiv Prepr arXiv1506.00019 (accessed on 13 May 2016).
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Yann Lecun Website. Available online: http://yann.lecun.com (accessed on 13 May 2016).
- Arenas, M.G.; Mora, A.M.; Romero, G.; Castillo, P.A. GPU Computation in bioinspired algorithms: A review. In Advances in Computational Intelligence; Springer: Berlin, Germany, 2011; pp. 433–440. [Google Scholar]
- Kirk, D.B.; Wen-Mei, W.H. Programming Massively Parallel Processors: A Hands-on Approach; Morgan Kaufmann: San Francisco, CA, USA, 2012. [Google Scholar]
- TOP 500 the List. Available online: http://top500.org (accessed on 13 May 2016).
- Stromatias, E.; Neil, D.; Pfeiffer, M.; Galluppi, F.; Furber, S.B.; Liu, S.-C. Robustness of spiking deep belief networks to noise and reduced bit precision of neuro-inspired hardware platforms. Front. Neurosci. 2015, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Izhikevich, E.M. Which model to use for cortical spiking neurons? IEEE Trans. Neural Netw. 2004, 15, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Kaggle. Available online: http://www.kaggle.com/c/MerckActivity (accessed on 13 May 2016).
- Ma, J.; Sheridan, R.P.; Liaw, A.; Dahl, G.E.; Svetnik, V. Deep neural nets as a method for quantitative structure–activity relationships. J. Chem. Inf. Model. 2015, 55, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Unterthiner, T.; Mayr, A.; Klambauer, G.; Steijaert, M.; Wegner, J.K.; Ceulemans, H.; Hochreiter, S. Deep learning as an opportunity in virtual screening. In Proceedings of the Deep Learning Workshop at NIPS, Montreal, QC, Canada, 8–13 December 2014.
- Unterthiner, T.; Mayr, A.; Klambauer, G.; Hochreiter, S. Toxicity Prediction Using Deep Learning. Available online: http://arXiv Prepr arXiv1503.01445 (accessed on 13 May 2016).
- Dahl, G.E. Deep Learning Approaches to Problems in Speech Recognition, Computational Chemistry, and Natural Language Text Processing. Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2015. [Google Scholar]
- Dahl, G.E.; Jaitly, N.; Salakhutdinov, R. Multi-Task Neural Networks for QSAR Predictions. Available online: http://arxiv.org/abs/1406.1231 (accessed on 13 May 2016).
- Ramsundar, B.; Kearnes, S.; Riley, P.; Webster, D.; Konerding, D.; Pande, V. Massively Multitask Networks for Drug Discovery. Available online: https://arxiv.org/abs/1502.02072 (accessed on 13 May 2016).
- Qi, Y.; Oja, M.; Weston, J.; Noble, W.S. A unified multitask architecture for predicting local protein properties. PLoS ONE 2012, 7, e32235. [Google Scholar] [CrossRef] [PubMed]
- Di Lena, P.; Nagata, K.; Baldi, P. Deep architectures for protein contact map prediction. Bioinformatics 2012, 28, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Eickholt, J.; Cheng, J. Predicting protein residue-residue contacts using deep networks and boosting. Bioinformatics 2012, 28, 3066–3072. [Google Scholar] [CrossRef] [PubMed]
- Eickholt, J.; Cheng, J. A study and benchmark of dncon: A method for protein residue-residue contact prediction using deep networks. BMC Bioinform. 2013, 14, S12. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.; Dehzangi, A.; Heffernan, R.; Sharma, A.; Paliwal, K.; Sattar, A.; Zhou, Y.; Yang, Y. Predicting backbone Cα angles and dihedrals from protein sequences by stacked sparse auto-encoder deep neural network. J. Comput. Chem. 2014, 35, 2040–2046. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, R.; Paliwal, K.; Lyons, J.; Dehzangi, A.; Sharma, A.; Wang, J.; Sattar, A.; Yang, Y.; Zhou, Y. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci. Rep. 2015, 5, 11476. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.P.; Shang, Y.; Xu, D. DL-PRO: A novel deep learning method for protein model quality assessment. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 6–11 July 2014; Volume 2014, pp. 2071–2078.
- Tan, J.; Ung, M.; Cheng, C.; Greene, C.S. Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders. Pac. Symp. Biocomput. 2014, 20, 132–143. [Google Scholar]
- Quang, D.; Chen, Y.; Xie, X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Wang, H.; Ganapathiraju, M. Learning structure in gene expression data using deep architectures, with an application to gene clustering. In Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1328–1335.
- Hubel, D.H.; Wiesel, T.N. Receptive fields and functional architecture of monkey striate cortex. J. Physiol. 1968, 195, 215–243. [Google Scholar] [CrossRef] [PubMed]
- Deep Learning. Available online: http://www.deeplearning.net/tutorial/lenet.html (accessed on 13 May 2016).
- Hughes, T.B.; Miller, G.P.; Swamidass, S.J. Modeling epoxidation of drug-like molecules with a deep machine learning network. ACS Cent. Sci. 2015, 1, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Guo, M.; Wang, C.; Liu, X.; Liu, Y.; Wu, X. MiRTDL: A deep learning approach for miRNA target prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015. [Google Scholar] [CrossRef] [PubMed]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Kellis, M. Deep learning for regulatory genomics. Nat. Biotechnol. 2015, 33, 825–826. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef]
- Graves, A.; Liwicki, M.; Fernández, S.; Bertolami, R.; Bunke, H.; Schmidhuber, J. A novel connectionist system for unconstrained handwriting recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 31, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Sak, H.; Senior, A.W.; Beaufays, F. Long short-term memory recurrent neural network architectures for large scale acoustic modeling. In Proceedings of the 2014 Interspeech, Carson City, NV, USA, 5–10 December 2013; pp. 338–342.
- Pascanu, R.; Gulcehre, C.; Cho, K.; Bengio, Y. How to construct deep recurrent neural networks. Available online: http://arXiv Prepr arXiv1312.6026 (accessed on 13 May 2016).
- Hermans, M.; Schrauwen, B. Training and analysing deep recurrent neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Carson City, NV, USA, 5–10 December 2013; pp. 190–198.
- Lusci, A.; Pollastri, G.; Baldi, P. Deep architectures and deep learning in chemoinformatics: The prediction of aqueous solubility for drug-like molecules. J. Chem. Inf. Model. 2013, 53, 1563–1575. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Dai, Z.; Chen, F.; Gao, S.; Pei, J.; Lai, L. Deep learning for drug-induced liver injury. J. Chem. Inf. Model. 2015, 55, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Sønderby, S.K.; Nielsen, H.; Sønderby, C.K.; Winther, O. Convolutional LSTM networks for subcellular localization of proteins. In Proceedings of the First Annual Danish Bioinformatics Conference, Odense, Denmark, 27–28 August 2015.
- Akopyan, F.; Sawada, J.; Cassidy, A.; Alvarez-Icaza, R.; Arthur, J.; Merolla, P.; Imam, N.; Nakamura, Y.; Datta, P.; Nam, G.-J. TrueNorth: Design and tool flow of a 65 mW 1 million neuron programmable neurosynaptic chip. IEEE Trans. Comput. Des. Integr. Circuits Syst. 2015, 34, 1537–1557. [Google Scholar] [CrossRef]
- Guo, X.; Ipek, E.; Soyata, T. Resistive computation: Avoiding the power wall with low-leakage, STT-MRAM based computing. ACM SIGARCH Comput. Archit. News. 2010, 38, 371–382. [Google Scholar] [CrossRef]
- McKee, S.A. Reflections on the memory wall. In Proceedings of the 1st conference on Computing Frontiers, Ischia, Italy, 14–16 April 2004; p. 162.
- Boncz, P.A.; Kersten, M.L.; Manegold, S. Breaking the memory wall in monetDB. Commun. ACM 2008, 51, 77–85. [Google Scholar] [CrossRef]
- Naylor, M.; Fox, P.J.; Markettos, A.T.; Moore, S.W. Managing the FPGA memory wall: Custom computing or vector processing? In Proceedings of the 2013 23rd International Conference on Field Programmable Logic and Applications (FPL), Porto, Portugal, 2–4 September 2013; pp. 1–6.
- Wen, W.; Wu, C.; Wang, Y.; Nixon, K.; Wu, Q.; Barnell, M.; Li, H.; Chen, Y. A New Learning Method for Inference Accuracy, Core Occupation, and Performance Co-Optimization on Truenorth Chip. 2016. Available online: http://arxiv.org/abs/1604.00697 (accessed on 13 May 2016).
- Mead, C.; Conway, L. Introduction to VLSI Systems; Addison-Wesley: Reading, MA, USA, 1980; Volume 1080. [Google Scholar]
- Esser, S.K.; Merolla, P.A.; Arthur, J.V.; Cassidy, A.S.; Appuswamy, R.; Andreopoulos, A.; Berg, D.J.; McKinstry, J.L.; Melano, T.; Barch, D.R.; et al. Convolutional Networks for Fast, Energy-Efficient Neuromorphic Computing. Available online: https://arxiv.org/abs/1603.08270 (accessed on 13 May 2016).
- Merolla, P.A.; Arthur, J.V.; Alvarez-Icaza, R.; Cassidy, A.S.; Sawada, J.; Akopyan, F.; Jackson, B.L.; Imam, N.; Guo, C.; Nakamura, Y.; et al. A million spiking-neuron integrated circuit with a scalable communication network and interface. Science 2014, 345, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Furber, S.B.; Galluppi, F.; Temple, S.; Plana, L. The SpiNNaker Project. Proc. IEEE 2014, 102, 652–665. [Google Scholar] [CrossRef]
- Dehaene, S. Consciousness and the Brain: Deciphering How the Brain Codes Our Thoughts; Viking Press: New York, NY, USA, 2014. [Google Scholar]
- Schemmel, J.; Brüderle, D.; Grübl, A.; Hock, M.; Meier, K.; Millner, S. A wafer-scale neuromorphic hardware system for large-scale neural modeling. In Proceedings of the ISCAS 2010—2010 IEEE International Symposium on Circuits and Systems: Nano-Bio Circuit Fabrics and Systems, Paris, France, 30 May–2 June 2010; pp. 1947–1950.
- Benjamin, B.V.; Gao, P.; McQuinn, E.; Choudhary, S.; Chandrasekaran, A.R.; Bussat, J.M.; Alvarez-Icaza, R.; Arthur, J.V.; Merolla, P.A.; Boahen, K. Neurogrid: A mixed-analog-digital multichip system for large-scale neural simulations. Proc. IEEE 2014, 102, 699–716. [Google Scholar] [CrossRef]
- Pastur-Romay, L.A.; Cedrón, F.; Pazos, A.; Porto-Pazos, A.B. Parallel Computation for Brain Simulation. Curr. Top. Med. Chem. Available online: https://www.researchgate.net/publication/284184342_Parallel_computation_for_Brain_Simulation (accessed on 5 August 2016).
- Pastur-Romay, L.A.; Cedrón, F.; Pazos, A.; Porto-Pazos, A.B. Computational models of the brain. In Proceedings of the MOL2NET International Conference on Multidisciplinary Sciences, Leioa, Spain, 5–15 December 2015.
- Amir, A.; Datta, P.; Risk, W.P.; Cassidy, A.S.; Kusnitz, J.A.; Esser, S.K.; Andreopoulos, A.; Wong, T.M.; Flickner, M.; Alvarez-Icaza, R.; et al. Cognitive computing programming paradigm: A corelet language for composing networks of neurosynaptic cores. In Proceedings of the International Joint Conference on Neural Networks, Dallas, TX, USA, 4–9 August 2013; pp. 1–10.
- Cassidy, A.S.; Alvarez-Icaza, R.; Akopyan, F.; Sawada, J.; Arthur, J.V.; Merolla, P.A.; Datta, P.; Tallada, M.G.; Taba, B.; Andreopoulos, A.; et al. Real-time scalable cortical computing at 46 giga-synaptic OPS/watt with ~100× speedup in time-to-solution and ~100,000× reduction in energy-to-solution. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, New Orleans, LA, USA, 16–21 November 2014; pp. 27–38.
- IBM. Available online: http://www.research.ibm.com/articles/brain-chips.shtml (accessed on 13 May 2016).
- Merolla, P.; Arthur, J.; Akopyan, F.; Imam, N.; Manohar, R.; Modha, D.S. A digital neurosynaptic core using embedded crossbar memory with 45pj per spike in 45nm. In Proceedings of the IEEE Custom Integrated Circuits Conference, San Jose, CA, USA, 19–21 September 2011; pp. 1–4.
- Sivilotti, M.A. Wiring considerations in analog VLSI systems, with application to field-programmable networks. Doctoral Dissertation, California Institute of Technology, Pasadena, CA, USA, 1990. [Google Scholar]
- Cabestany, J.; Prieto, A.; Sandoval, F. Computational Intelligence and Bioinspired Systems; Lecture Notes in Computer Science; Springer Berlin Heidelberg: Berlin & Heidelberg, Germany, 2005; Volume 3512. [Google Scholar]
- Preissl, R.; Wong, T.M.; Datta, P.; Flickner, M.D.; Singh, R.; Esser, S.K.; Risk, W.P.; Simon, H.D.; Modha, D.S. Compass: A scalable simulator for an architecture for cognitive computing. In Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, Slat Lake City, UT, USA, 11–15 November 2012; pp. 1–11.
- Minkovich, K.; Thibeault, C.M.; O’Brien, M.J.; Nogin, A.; Cho, Y.; Srinivasa, N. HRLSim: A high performance spiking neural network simulator for GPGPU clusters. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Ananthanarayanan, R.; Modha, D.S. Anatomy of a cortical simulator. In Proceedings of the 2007 ACM/IEEE Conference on Supercomputing, Reno, NV, USA, 10–16 November 2007.
- Modha, D.S.; Ananthanarayanan, R.; Esser, S.K.; Ndirango, A.; Sherbondy, A.J.; Singh, R. Cognitive Computing. Commun. ACM 2011, 54, 62. [Google Scholar] [CrossRef]
- Wong, T.M.; Preissl, R.; Datta, P.; Flickner, M.; Singh, R.; Esser, S.K.; Mcquinn, E.; Appuswamy, R.; Risk, W.P.; Simon, H.D.; et al. “1014” IBM Research Divsion, Research Report RJ10502, 2012. IBM J. Rep. 2012, 10502, 13–15. [Google Scholar]
- Cassidy, A.S.; Merolla, P.; Arthur, J.V.; Esser, S.K.; Jackson, B.; Alvarez-Icaza, R.; Datta, P.; Sawada, J.; Wong, T.M.; Feldman, V.; et al. Cognitive computing building block: A versatile and efficient digital neuron model for neurosynaptic cores. In Proceedings of the 2013 International Joint Conference on Neural Networks (IJCNN), Dallas, TX, USA, 4–9 August 2013; pp. 1–10.
- Esser, S.K.; Andreopoulos, A.; Appuswamy, R.; Datta, P.; Barch, D.; Amir, A.; Arthur, J.; Cassidy, A.; Flickner, M.; Merolla, P.; et al. Cognitive computing systems: Algorithms and applications for networks of neurosynaptic cores. In Proceedings of the 2013 International Joint Conference on Neural Networks (IJCNN), Dallas, TX, USA, 4–9 August 2013; pp. 1–10.
- Diehl, P.U.; Pedroni, B.U.; Cassidy, A.; Merolla, P.; Neftci, E.; Zarrella, G. TrueHappiness: Neuromorphic Emotion Recognition on Truenorth. Available online: http://arxiv.org/abs/1601.04183 (accessed on 13 May 2016).
- Navaridas, J.; Luján, M.; Plana, L.A.; Temple, S.; Furber, S.B. SpiNNaker: Enhanced multicast routing. Parallel Comput. 2015, 45, 49–66. [Google Scholar] [CrossRef]
- Furber, S.B.; Lester, D.R.; Plana, L.A.; Garside, J.D.; Painkras, E.; Temple, S.; Brown, A.D. Overview of the SpiNNaker system architecture. IEEE Trans. Comput. 2013, 62, 2454–2467. [Google Scholar] [CrossRef]
- Navaridas, J.; Luján, M.; Miguel-Alonso, J.; Plana, L.A.; Furber, S. Understanding the interconnection network of spiNNaker. In Proceedings of the 23rd international conference on Conference on Supercomputing—ICS ‘09, Yorktown Heights, NY, USA, 8–12 June 2009; pp. 286–295.
- Plana, L.; Furber, S.B.; Temple, S.; Khan, M.; Shi, Y.; Wu, J.; Yang, S. A GALS infrastructure for a massively parallel multiprocessor. Des. Test Comput. IEEE 2007, 24, 454–463. [Google Scholar] [CrossRef]
- Furber, S.; Brown, A. Biologically-inspired massively-parallel architectures- computing beyond a million processors. In Proceedings of the International Conference on Application of Concurrency to System Design, Augsburg, Germany, 1–3 July 2009; pp. 3–12.
- Davies, S.; Navaridas, J.; Galluppi, F.; Furber, S. Population-based routing in the spiNNaker neuromorphic architecture. In Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), Brisbane, Australia, 10–15 June 2012; pp. 1–8.
- Galluppi, F.; Lagorce, X.; Stromatias, E.; Pfeiffer, M.; Plana, L.A.; Furber, S.B.; Benosman, R.B. A framework for plasticity implementation on the spiNNaker neural architecture. Front. Neurosci. 2014, 8, 429. [Google Scholar] [CrossRef] [PubMed]
- Galluppi, F.; Davies, S.; Rast, A.; Sharp, T.; Plana, L.A.; Furber, S. A hierachical configuration system for a massively parallel neural hardware platform. In Proceedings of the 9th Conference on Computing Frontiers—CF ′12, Caligari, Italy, 15–17 May 2012; ACM Press: New York, NY, USA, 2012; p. 183. [Google Scholar]
- Davison, A.P. PyNN: A common interface for neuronal network simulators. Front. Neuroinform. 2008, 2. [Google Scholar] [CrossRef] [PubMed]
- Stewart, T.C.; Tripp, B.; Eliasmith, C. Python Scripting in the Nengo Simulator. Front. Neuroinform. 2009, 3. [Google Scholar] [CrossRef] [PubMed]
- Bekolay, T.; Bergstra, J.; Hunsberger, E.; DeWolf, T.; Stewart, T.C.; Rasmussen, D.; Choo, X.; Voelker, A.R.; Eliasmith, C. Nengo: A python tool for building large-scale functional brain models. Front. Neuroinform. 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.; Stewart, T.; Eliasmith, C.; Furber, S. Spike-based learning of transfer functions with the SpiNNaker neuromimetic simulator. In Proceedings of the 2013 International Joint Conference on Neural Networks (IJCNN), Dallas, TX, USA, 4–9 August 2013; pp. 1–8.
- Jin, X.; Luján, M.; Plana, L.A.; Rast, A.D.; Welbourne, S.R.; Furber, S.B. Efficient parallel implementation of multilayer backpropagation networks on spiNNaker. In Proceedings of the 7th ACM International Conference on Computing Frontiers, Bertinoro, Italy, 17–19 May 2010; pp. 89–90.
- Serrano- Gotarredona, T.; Linares-Barranco, B.; Galluppi, F.; Plana, L.; Furber, S. ConvNets Experiments on SpiNNaker. In 2015 IEEE International Symposium on Circuits and Systems (ISCAS); IEEE: Piscataway, NJ, USA, 2015; pp. 2405–2408. [Google Scholar]
- Trask, A.; Gilmore, D.; Russell, M. Modeling order in neural word embeddings at scale. In Proceedings of the 32nd International Conference on Machine Learning (ICML-15), Lille, France, 6–11 July 2015.
- Smith, E.C.; Lewicki, M.S. Efficient auditory coding. Nature 2006, 439, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Kurzweil, R. The Singularity Is Near: When Humans Transcend Biology; Viking: New York, NY, USA, 2005. [Google Scholar]
- Kurzweil, R. The Law of Accelerating Returns; Springer: Berlin, Germany, 2004. [Google Scholar]
- Von Neumann, J.; Kurzweil, R. The Computer and the Brain; Yale University Press: New Haven, CT, USA, 2012. [Google Scholar]
- Kurzweil, R. How to Create a Mind: The Secret of Human Thought Revealed; Viking: New York, NY, USA, 2012; Volume 2012. [Google Scholar]
- Merkle, R. How Many Bytes in Human Memory? Foresight Update: Palo Alto, CA, USA, 1988; Volume 4. [Google Scholar]
- Merkle, R.C. Energy Limits to the Computational Power of the Human Brain; Foresight Update: Palo Alto, CA, USA, 1989; Volume 6. [Google Scholar]
- Deep Learning Blog. Available online: http://timdettmers.com/2015/07/27/brain-vs-deep-learning-singularity/ (accessed on 13 May 2016).
- Azevedo, F.A.C.; Carvalho, L.R.B.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.L.; Leite, R.E.P.; Jacob Filho, W.; Lent, R.; Herculano-Houzel, S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009, 513, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D. The Other Brain: From Dementia to Schizophrenia, How New Discoveries about the Brain Are Revolutionizing Medicine and Science; Simon and Schuster: New York City, NY, USA, 2009. [Google Scholar]
- Koob, A. The Root of Thought: Unlocking Glia—The Brain Cell That Will Help Us Sharpen Our Wits, Heal Injury, and Treat Brain Disease; FT Press: Upper Saddle River, NJ, USA, 2009. [Google Scholar]
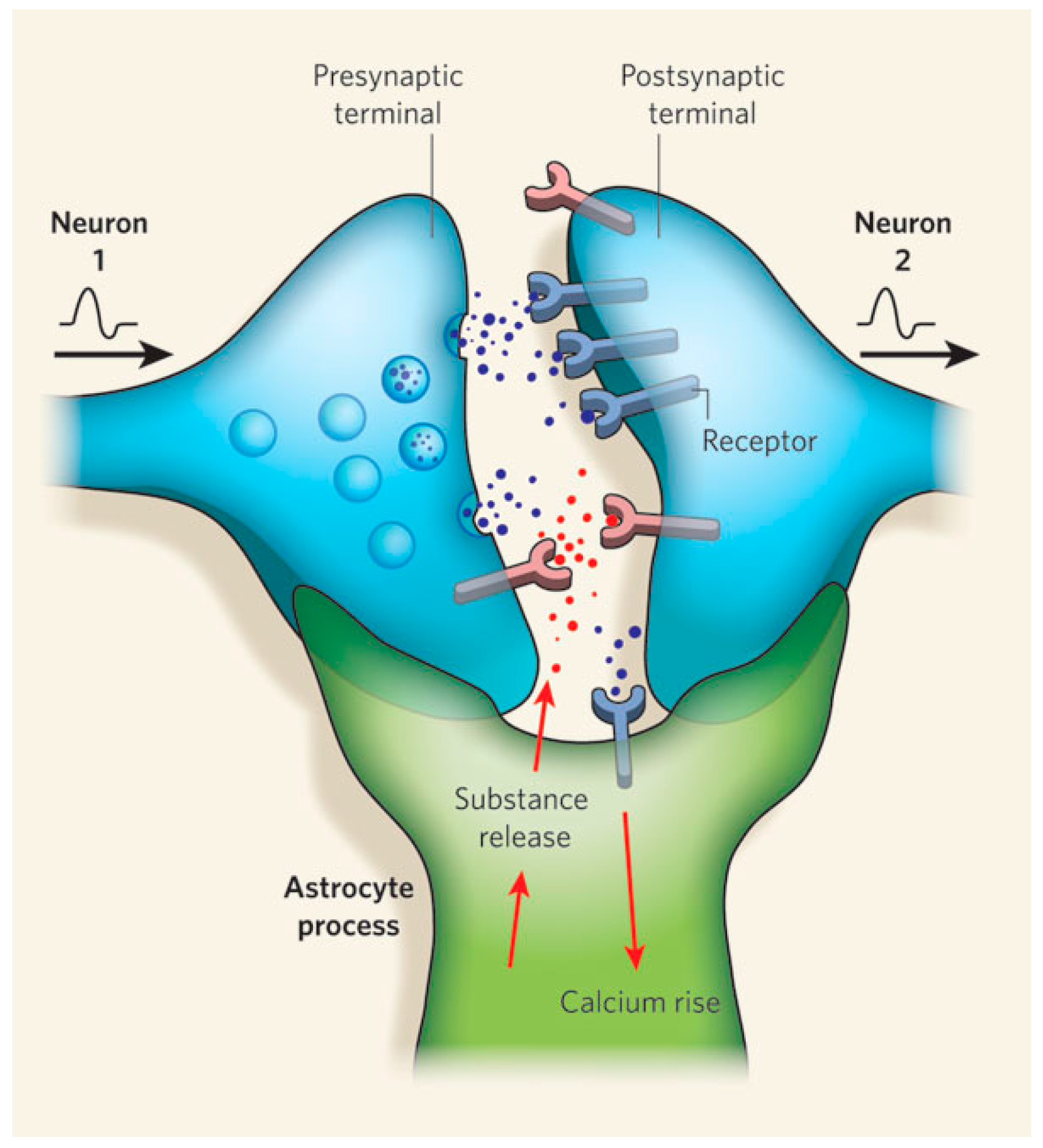
- Fields, R.; Araque, A.; Johansen-Berg, H. Glial biology in learning and cognition. Neuroscientist 2013, 20, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Perea, G.; Sur, M.; Araque, A. Neuron-glia networks: Integral gear of brain function. Front. Cell Neurosci. 2014, 8, 378. [Google Scholar] [CrossRef] [PubMed]
- Wade, J.J.; McDaid, L.J.; Harkin, J.; Crunelli, V.; Kelso, J.A. Bidirectional coupling between astrocytes and neurons mediates learning and dynamic coordination in the brain: A multiple modeling approach. PLoS ONE 2011, 6, e29445. [Google Scholar] [CrossRef] [PubMed]
- Haydon, P.G.; Nedergaard, M. How do astrocytes participate in neural plasticity? Cold Spring Harb. Perspect. Biol. 2015, 7, a020438. [Google Scholar] [CrossRef] [PubMed]
- Araque, A.; Parpura, V.; Sanzgiri, R.P.; Haydon, P.G. Tripartite synapses: Glia, the unacknowledged partner. Trends Neurosci. 1999, 22, 208–215. [Google Scholar] [CrossRef]
- Allen, N.J.; Barres, B.A. Neuroscience: Glia—More than Just Brain Glue. Nature 2009, 457, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Zorec, R.; Araque, A.; Carmignoto, G.; Haydon, P.G.; Verkhratsky, A.; Parpura, V. Astroglial excitability and gliotransmission: An appraisal of Ca2+ as a signalling route. ASN Neuro 2012. [Google Scholar] [CrossRef] [PubMed]
- Araque, A.; Carmignoto, G.; Haydon, P.G.; Oliet, S.H.R.; Robitaille, R.; Volterra, A. Gliotransmitters travel in time and space. Neuron 2014, 81, 728–739. [Google Scholar] [CrossRef] [PubMed]
- Ben Achour, S.; Pascual, O. Glia: The many ways to modulate synaptic plasticity. Neurochem. Int. 2010, 57, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Schafer, D.P.; Lehrman, E.K.; Stevens, B. The “quad-partite” synapse: Microglia-synapse interactions in the developing and mature CNS. Glia 2013, 61, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Oberheim, N.A.; Goldman, S.A.; Nedergaard, M. Heterogeneity of astrocytic form and function. Methods Mol. Biol. 2012, 814, 23–45. [Google Scholar] [PubMed]
- Oberheim, N.A.; Takano, T.; Han, X.; He, W.; Lin, J.H.C.; Wang, F.; Xu, Q.; Wyatt, J.D.; Pilcher, W.; Ojemann, J.G.; et al. Uniquely hominid features of adult human astrocytes. J. Neurosci. 2009, 29, 3276–3287. [Google Scholar] [CrossRef] [PubMed]
- Nedergaard, M.; Ransom, B.; Goldman, S.A. New roles for astrocytes: Redefining the functional architecture of the brain. Trends Neurosci. 2003, 26, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, C.C.; Stimpson, C.D.; Raghanti, M.A.; Wildman, D.E.; Uddin, M.; Grossman, L.I.; Goodman, M.; Redmond, J.C.; Bonar, C.J.; Erwin, J.M.; et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc. Natl. Acad. Sci. USA 2006, 103, 13606–13611. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.; Parker, A.C.; Tseng, K. An in-silico glial microdomain to invoke excitability in cortical neural networks. In Proceedings of the 2011 IEEE International Symposium of Circuits and Systems (ISCAS), Rio de Janeiro, Brazil, 15–18 May 2011; pp. 681–684.
- Irizarry-Valle, Y.; Parker, A.C.; Joshi, J. A CMOS neuromorphic approach to emulate neuro-astrocyte interactions. In Proceedings of the International Joint Conference on Neural Networks, Dallas, TX, USA, 4–9 August 2013.
- Irizarry-Valle, Y.; Parker, A.C. Astrocyte on neuronal phase synchrony in CMOS. In Proceedings of the 2014 IEEE International Symposium on Circuits and Systems (ISCAS), Melbourne, Australia, 1–5 June 2014; pp. 261–264.
- Irizarry-Valle, Y.; Parker, A.C. An astrocyte neuromorphic circuit that influences neuronal phase synchrony. IEEE Trans. Biomed. Circuits Syst. 2015, 9, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Nazari, S.; Amiri, M.; Faez, K.; Amiri, M. Multiplier-less digital implementation of neuron-astrocyte signalling on FPGA. Neurocomputing 2015, 164, 281–292. [Google Scholar] [CrossRef]
- Nazari, S.; Faez, K.; Amiri, M.; Karami, E. A digital implementation of neuron–astrocyte interaction for neuromorphic applications. Neural Netw. 2015, 66, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Nazari, S.; Faez, K.; Karami, E.; Amiri, M. A digital neurmorphic circuit for a simplified model of astrocyte dynamics. Neurosci. Lett. 2014, 582, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Porto, A.; Pazos, A.; Araque, A. Artificial neural networks based on brain circuits behaviour and genetic algorithms. In Computational Intelligence and Bioinspired Systems; Springer: Berlin, Germany, 2005; pp. 99–106. [Google Scholar]
- Porto, A.; Araque, A.; Rabuñal, J.; Dorado, J.; Pazos, A. A new hybrid evolutionary mechanism based on unsupervised learning for connectionist systems. Neurocomputing 2007, 70, 2799–2808. [Google Scholar] [CrossRef]
- Porto-Pazos, A.B.; Veiguela, N.; Mesejo, P.; Navarrete, M.; Alvarellos, A.; Ibáñez, O.; Pazos, A.; Araque, A. Artificial astrocytes improve neural network performance. PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed]
- Alvarellos-González, A.; Pazos, A.; Porto-Pazos, A.B. Computational models of neuron-astrocyte interactions lead to improved efficacy in the performance of neural networks. Comput. Math. Methods Med. 2012. [Google Scholar] [CrossRef] [PubMed]
- Mesejo, P.; Ibáñez, O.; Fernández-Blanco, E.; Cedrón, F.; Pazos, A.; Porto-Pazos, A.B. Artificial neuron–glia networks learning approach based on cooperative coevolution. Int. J. Neural Syst. 2015, 25, 1550012. [Google Scholar] [CrossRef] [PubMed]
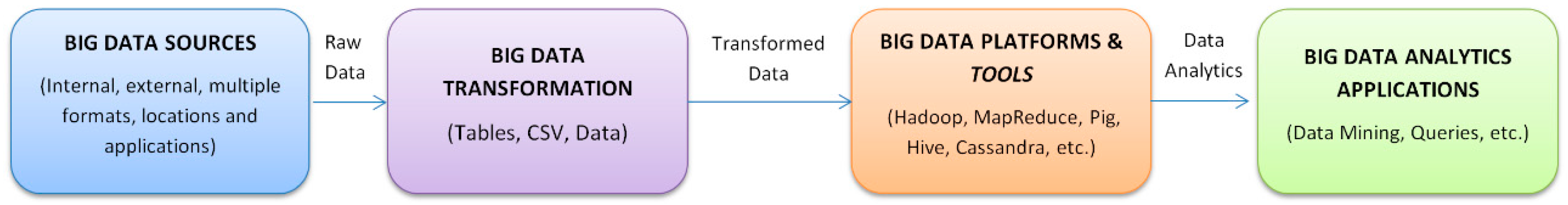
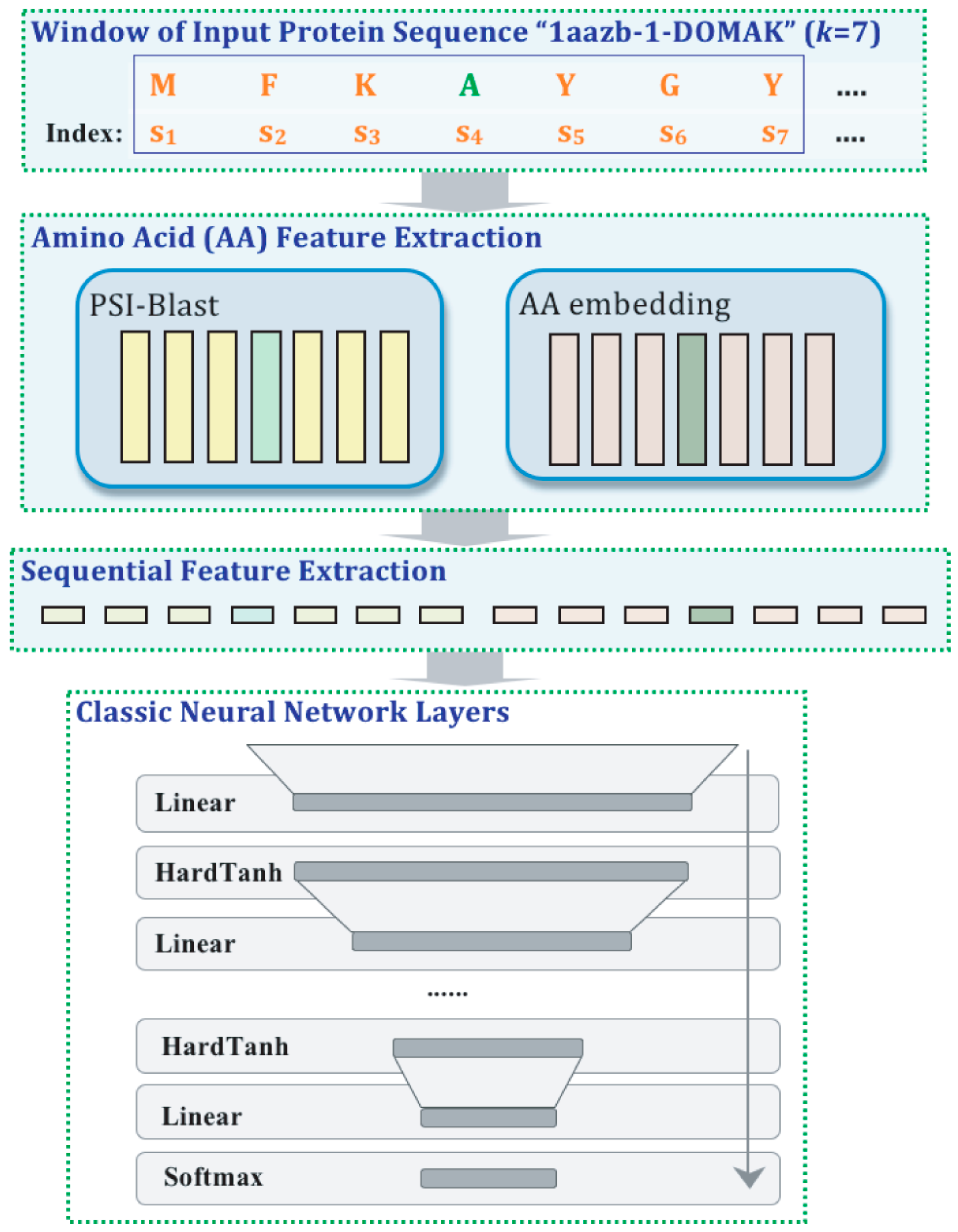

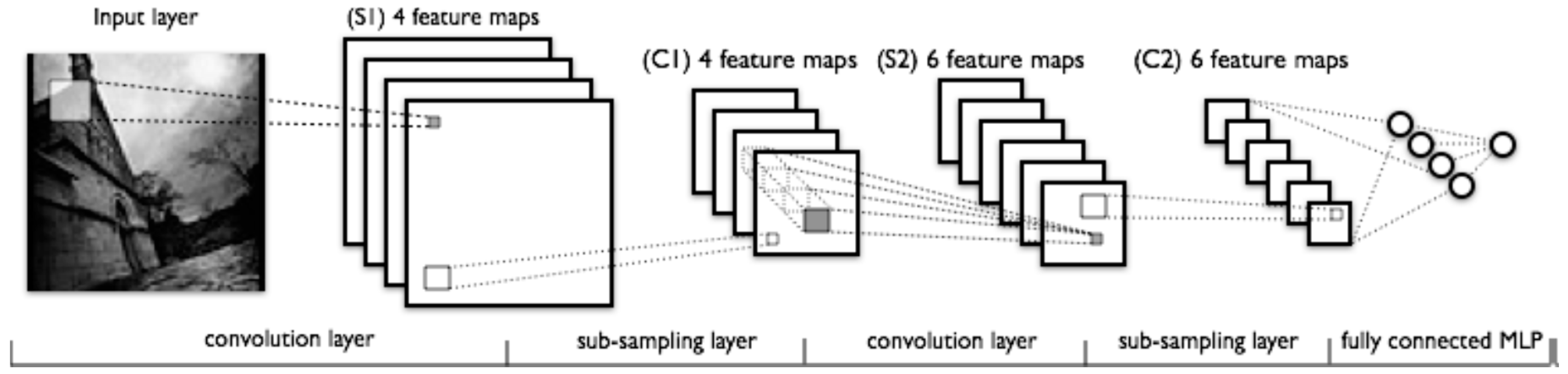

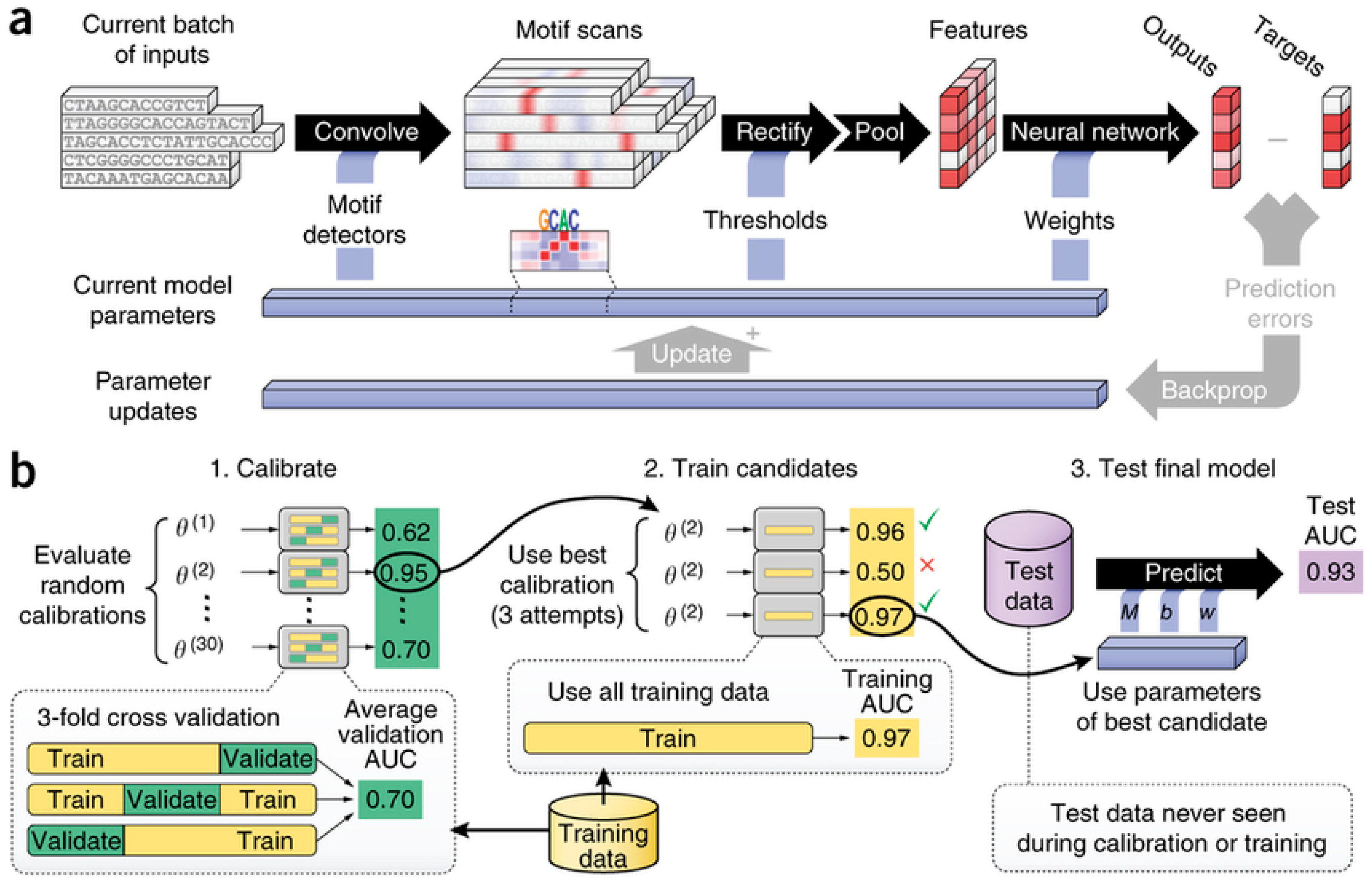

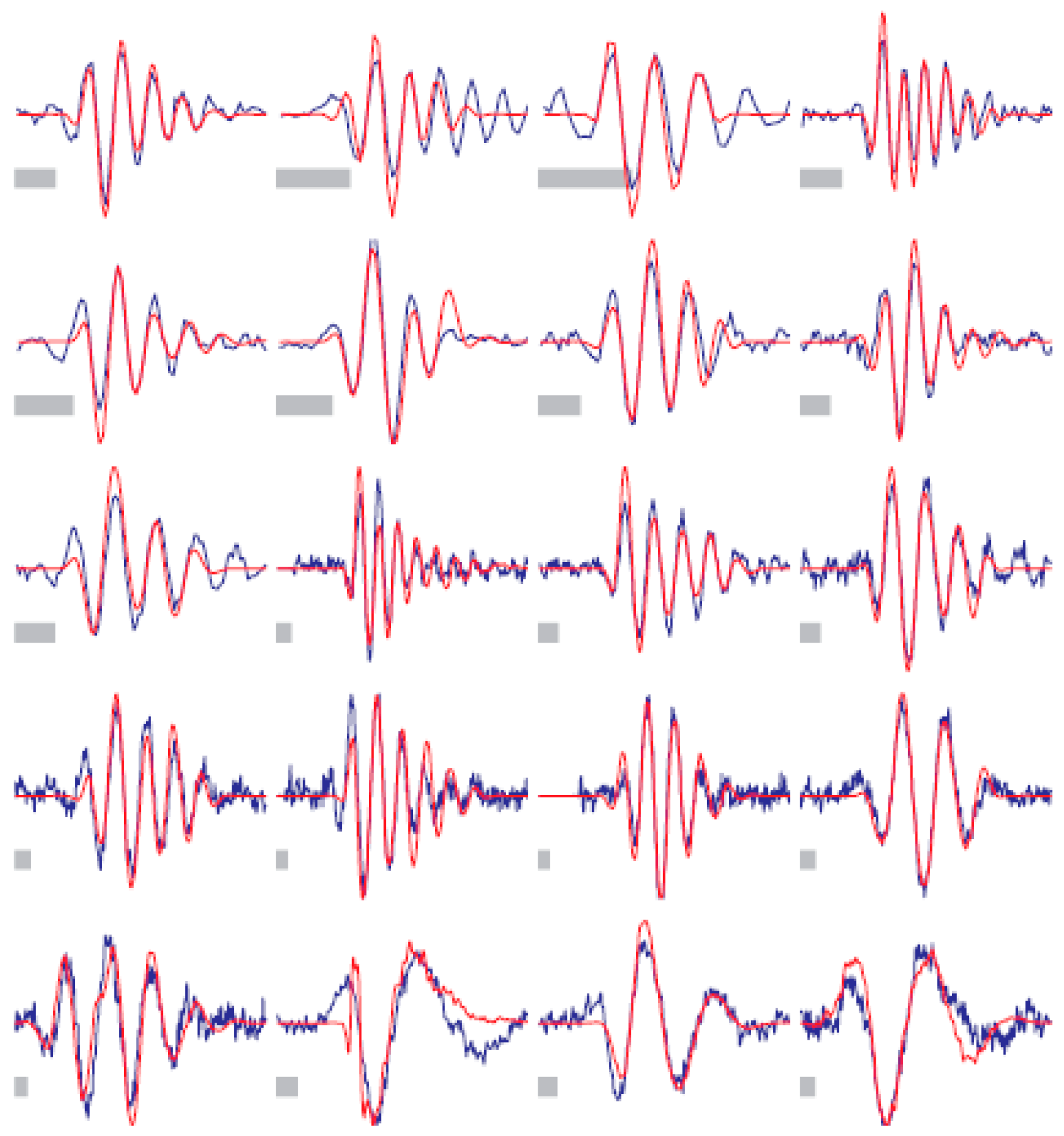
| Task (Year) | Competition |
|---|---|
| Handwriting recognition (2009) | MNIST (many), Arabic HWX (IDSIA) |
| Volumetric brain image segmentation (2009) | Connectomics (IDSIA, MIT) |
| OCR in the Wild (2011) | StreetView House Numbers (NYU and others) |
| Traffic sign recognition (2011) | GTSRB competition (IDSIA, NYU) |
| Human Action Recognition (2011) | Hollywood II dataset (Stanford) |
| Breast cancer cell mitosis detection (2011) | MITOS (IDSIA) |
| Object Recognition (2012) | ImageNet competition (Toronto) |
| Scene Parsing (2012) | Stanford bgd, SiftFlow, Barcelona datasets (NYU) |
| Speech Recognition (2012) | Acoustic modeling (IBM and Google) |
| Asian handwriting recognition (2013) | ICDAR competition (IDSIA) |
| Pedestrian Detection (2013) | INRIA datasets and others (NYU) |
| Scene parsing from depth images (2013) | NYU RGB-D dataset (NYU) |
| Playing Atari games (2013) | 2600 Atari games (Google DeepMind Technologies) |
| Game of Go (2016) | AlphaGo vs. Human World Champion (Google DeepMind Technologies) |
| Network Architecture | Pharmacology | Bioinformatics |
|---|---|---|
| DAEN | [1,2,3,4,5,6,7,23] | [8,9,10,11,12,13,14,15,16,17] |
| DCNN | [18] | [19,20,21] |
| DRNN | [22,23] | [24] |
| Method | AUC | p-Value |
|---|---|---|
| Deep Auto-Encoder Network | 0.830 | – |
| Support Vector Machine | 0.816 | 1.0 × 10−7 |
| Binary Kernel Discrimination | 0.803 | 1.9 × 10−67 |
| Logistic Regression | 0.796 | 6.0 × 10−53 |
| k-Nearest neighbor | 0.775 | 2.5 × 10−142 |
| Pipeline Pilot Bayesian Classifier | 0.755 | 5.4 × 10−116 |
| Parzen-Rosenblatt | 0.730 | 1.8 × 10−153 |
| Similarity Ensemble Approach | 0.699 | 1.8 × 10−173 |
| Article Identifier | Assay Target/Goal | Assay Type | #Active | #Inactive |
|---|---|---|---|---|
| 1851(2c19) | Cytochrome P450, family 2, subfamily C, polypeptide 19 | Biochemical | 5913 | 7532 |
| 1851(2d6) | Cytochrome P450, family 2, subfamily D, polypeptide 6, isoform 2 | Biochemical | 2771 | 11,139 |
| 1851(3a4) | Cytochrome P450, family 3, subfamily A, polypeptide 14 | Biochemical | 5266 | 7751 |
| 1851(1a2) | Cytochrome P450, family 1, subfamily A, polypeptide 2 | Biochemical | 6000 | 7256 |
| 1851(2c9) | Cytochrome P450, family 2, subfamily C, polypeptide 9 | Biochemical | 4119 | 8782 |
| 1915 | Group A Streptokinase Expression Inhibition | Cell | 2219 | 1017 |
| 2358 | Protein phosphatase 1, catalytic subunit, α isoform 3 | Biochemical | 1006 | 934 |
| 463213 | Identify small molecule inhibitors of tim10-1 yeast | Cell | 4141 | 3235 |
| 463215 | Identify small molecule inhibitors of tim10 yeast | Cell | 2941 | 1695 |
| 488912 | Identify inhibitors of Sentrin-specific protease 8 (SENP8) | Biochemical | 2491 | 3705 |
| 488915 | Identify inhibitors of Sentrin-specific protease 6 (SENP6) | Biochemical | 3568 | 2628 |
| 488917 | Identify inhibitors of Sentrin-specific protease 7 (SENP7) | Biochemical | 4283 | 1913 |
| 488918 | Identify inhibitors of Sentrin-specific proteases (SENPs) using a Caspase-3 Selectivity assay | Biochemical | 3691 | 2505 |
| 492992 | Identify inhibitors of the two-pore domain potassium channel (KCNK9) | Cell | 2094 | 2820 |
| 504607 | Identify inhibitors of Mdm2/MdmX interaction | Cell | 4830 | 1412 |
| 624504 | Inhibitor hits of the mitochondrial permeability transition pore | Cell | 3944 | 1090 |
| 651739 | Inhibition of Trypanosoma cruzi | Cell | 4051 | 1324 |
| 615744 | NIH/3T3 (mouse embryonic fibroblast) toxicity | Cell | 3102 | 2306 |
| 652065 | Identify molecules that bind r (CAG) RNA repeats | Cell | 2966 | 1287 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pastur-Romay, L.A.; Cedrón, F.; Pazos, A.; Porto-Pazos, A.B. Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications. Int. J. Mol. Sci. 2016, 17, 1313. https://doi.org/10.3390/ijms17081313
Pastur-Romay LA, Cedrón F, Pazos A, Porto-Pazos AB. Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications. International Journal of Molecular Sciences. 2016; 17(8):1313. https://doi.org/10.3390/ijms17081313
Chicago/Turabian StylePastur-Romay, Lucas Antón, Francisco Cedrón, Alejandro Pazos, and Ana Belén Porto-Pazos. 2016. "Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications" International Journal of Molecular Sciences 17, no. 8: 1313. https://doi.org/10.3390/ijms17081313
APA StylePastur-Romay, L. A., Cedrón, F., Pazos, A., & Porto-Pazos, A. B. (2016). Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications. International Journal of Molecular Sciences, 17(8), 1313. https://doi.org/10.3390/ijms17081313








