RETRACTED: 6-Pyrazolylpurine as an Artificial Nucleobase for Metal-Mediated Base Pairing in DNA Duplexes
Abstract
:1. Introduction
2. Results
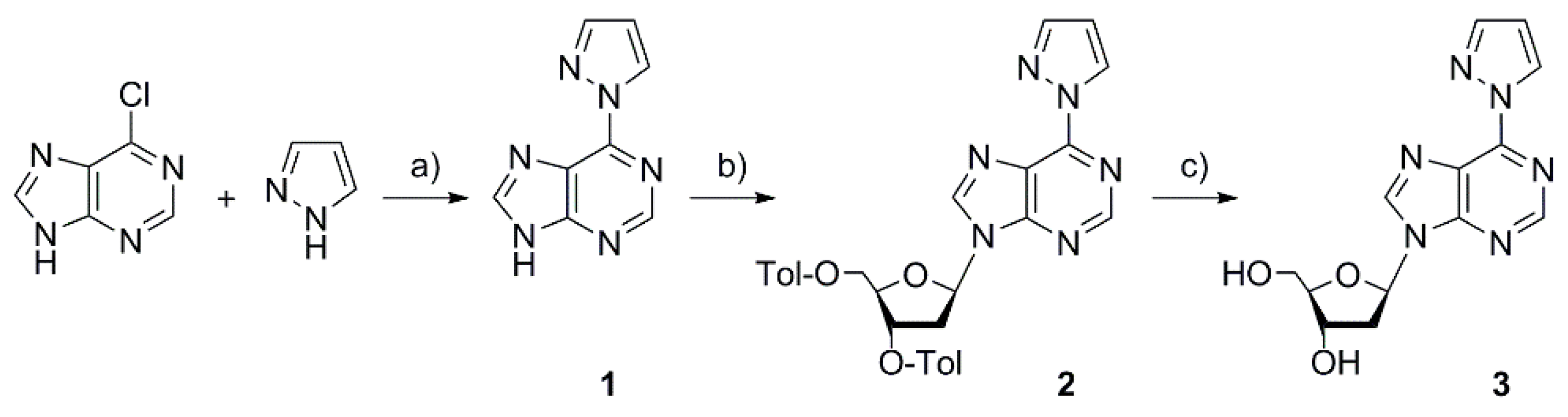
2.1. Synthesis of the Deoxyribonucleoside and the DNA Duplex
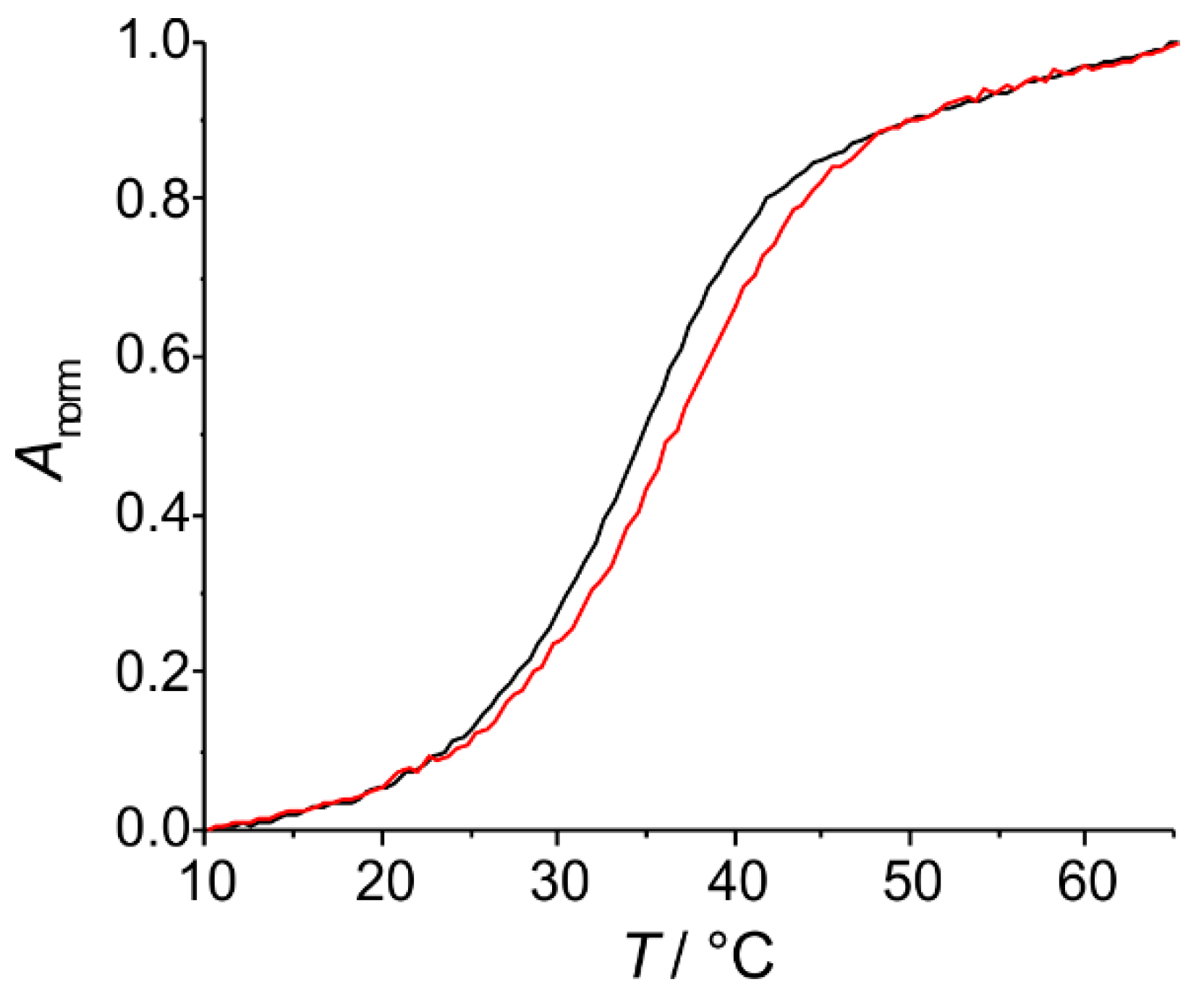
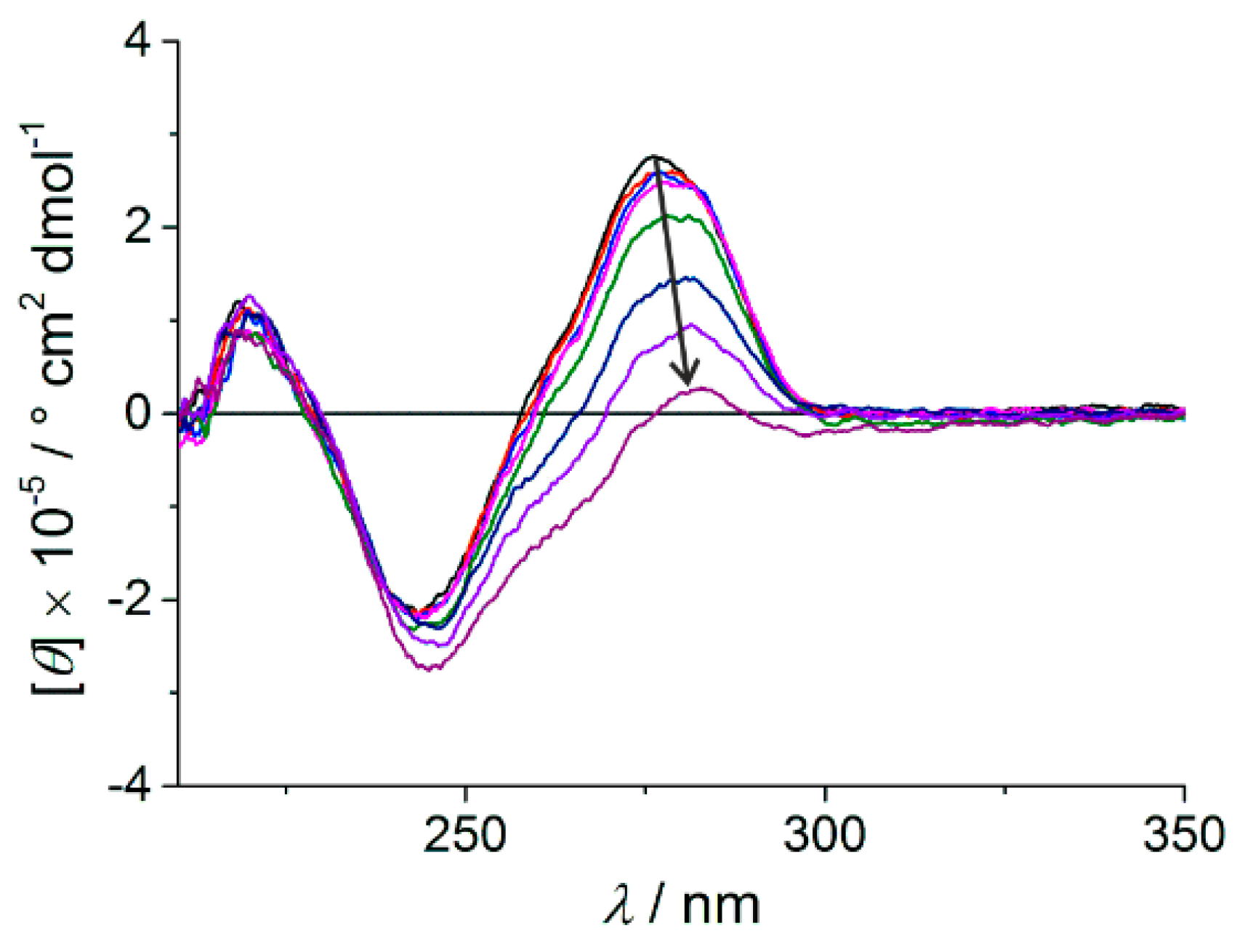
2.2. Spectroscopic Characterization of the Metal-Containing DNA Duplex
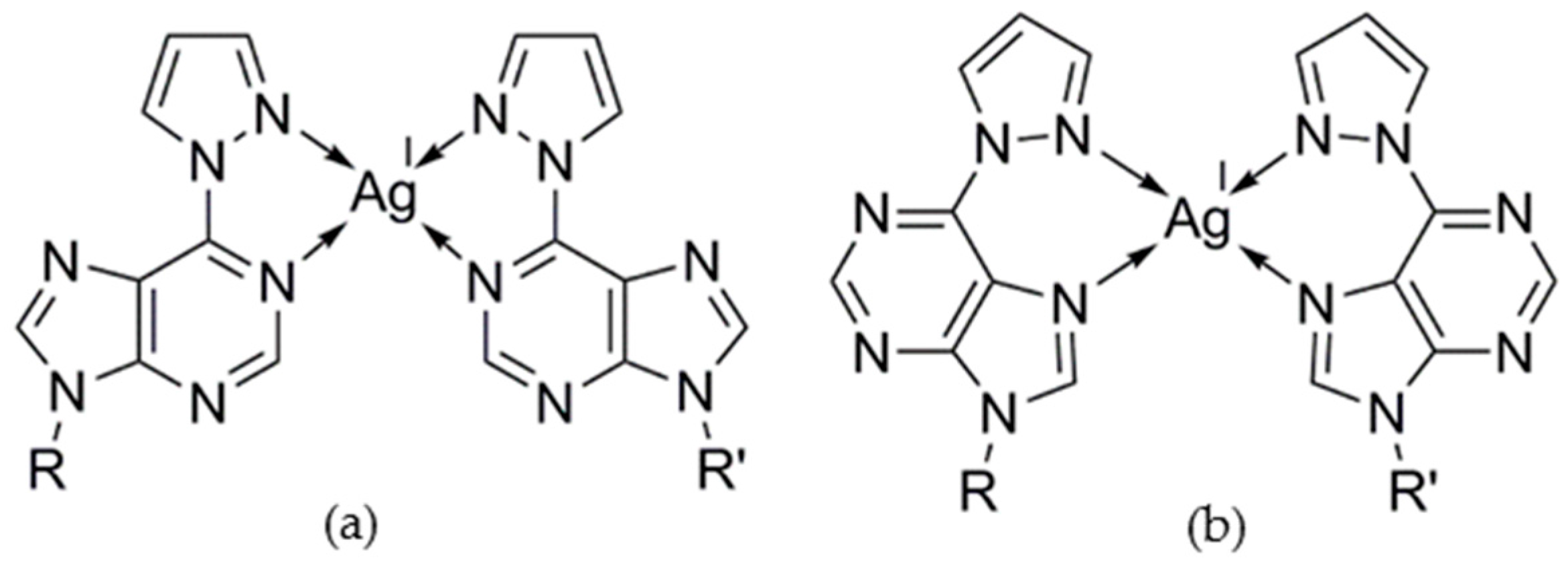
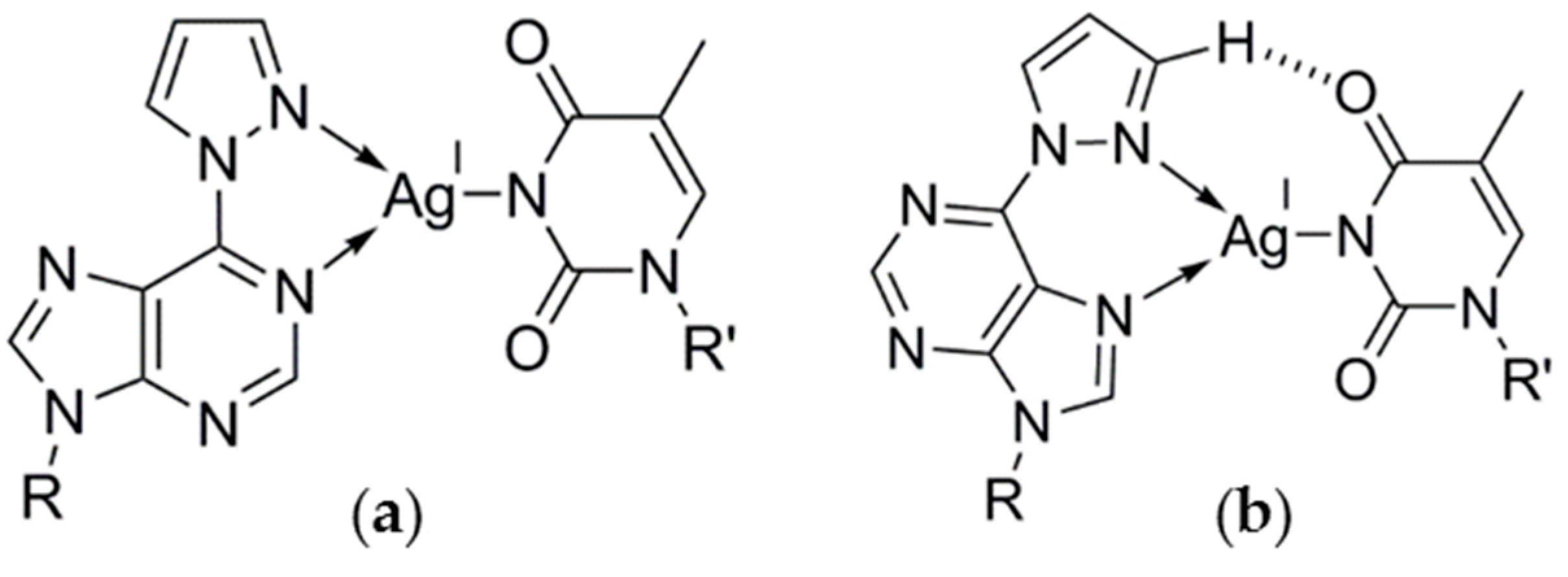
2.2.1. Homo Base Pair (6PP:6PP)
2.2.2. Hetero Base Pairs (6PP:C and 6PP:T)
3. Discussion
4. Materials and Methods
4.1. Oligonucleotide Synthesis and Characterization
4.2. Spectroscopy
4.3. 6-(1H-Pyrazol-1-yl)-9H-purine 1
4.4. 9-(2-Deoxy-3,5-di-O-p-toluoyl-β-d-erythro-pentofuranosyl)-6-(1H-pyrazol-1-yl)-9H-purine 2
4.5. 9-(2-Deoxy-β-d-erythro-pentofuranosyl)-6-(1H-pyrazol-1-yl)-9H-purine 3
4.6. 9-[2-Deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-6-(1H-pyrazol-1-yl)-9H-purine 4
4.7. 9-[2-Deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-6-(1H-pyrazol-1-yl)-9H-purine 3′-(2-cyanoethyl)-N,N′-diisopropyl phosphoramidite 5
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 6PP | 6-Pyrazol-1-yl-purine |
| A | Adenine |
| C | Cytosine |
| CD | Circular dichroism |
| G | Guanine |
| MOPS | 3-(N-Morpholino) propane sulfonate |
| T | Thymine |
References
- Stulz, E.; Clever, G.H. DNA in Supramolecular Chemistry and Nanotechnology; John Wiley & Sons, Ltd.: Chichester, UK, 2015. [Google Scholar]
- Müller, J. Metal-ion-mediated base pairs in nucleic acids. Eur. J. Inorg. Chem. 2008. [Google Scholar] [CrossRef]
- Takezawa, Y.; Shionoya, M. Metal-mediated DNA base pairing: Alternatives to hydrogen-bonded Watson–Crick base pairs. Acc. Chem. Res. 2012, 45, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kondo, J.; Sychrovský, V.; Šebera, J.; Dairaku, T.; Saneyoshi, H.; Urata, H.; Torigoe, H.; Ono, A. Structures, physicochemical properties, and applications of T–HgII–T, C–AgI–C, and other metallo-base-pairs. Chem. Commun. 2015, 51, 17343–17360. [Google Scholar] [CrossRef] [PubMed]
- Clever, G.H.; Kaul, C.; Carell, T. DNA-metal base pairs. Angew. Chem. Int. Ed. 2007, 46, 6226–6236. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, S.; Megger, N.; Böhme, D.; Sigel, R.K.O.; Müller, J. Solution structure of a DNA double helix with consecutive metal-mediated base pairs. Nat. Chem. 2010, 2, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Kumbhar, S.; Johannsen, S.; Sigel, R.K.O.; Waller, M.P.; Müller, J. A QM/MM refinement of an experimental DNA structure with metal-mediated base pairs. J. Inorg. Biochem. 2013, 127, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Yamada, T.; Hirose, C.; Okamoto, I.; Tanaka, Y.; Ono, A. Crystal structure of metallo-DNA duplex containing consecutive Watson–Crick-like T–HgII–T base pairs. Angew. Chem. Int. Ed. 2014, 53, 2385–2388. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Meggers, E.; Spraggon, G.; Schultz, P.G. Structure of a copper-mediated base pair in DNA. J. Am. Chem. Soc. 2001, 123, 12364–12367. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Tada, Y.; Dairaku, T.; Saneyoshi, H.; Okamoto, I.; Tanaka, Y.; Ono, A. High-resolution crystal structure of a silver(I)–RNA hybrid duplex containing Watson–Crick-like C-silver(I)-C metallo-base pairs. Angew. Chem. Int. Ed. 2015, 54, 13323–13326. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, S.; Paulus, S.; Düpre, N.; Müller, J.; Sigel, R.K.O. Using in vitro transcription to construct scaffolds for one-dimensional arrays of mercuric ions. J. Inorg. Biochem. 2008, 102, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Scharf, P.; Müller, J. Nucleic acids with metal-mediated base pairs and their applications. ChemPlusChem 2013, 78, 20–34. [Google Scholar] [CrossRef]
- Kaul, C.; Müller, M.; Wagner, M.; Schneider, S.; Carell, T. Reversible bond formation enables the replication and amplification of a crosslinking salen complex as an orthogonal base pair. Nat. Chem. 2011, 3, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Clever, G.H.; Takezawa, Y.; Kaneko, M.; Tanaka, K.; Guo, X.; Shionoya, M. Direct conductance measurement of individual metallo-DNA duplexes within single-molecule break junctions. Angew. Chem. Int. Ed. 2011, 50, 8886–8890. [Google Scholar] [CrossRef] [PubMed]
- Ehrenschwender, T.; Schmucker, W.; Wellner, C.; Augenstein, T.; Carl, P.; Harmer, J.; Breher, F.; Wagenknecht, H.-A. Development of a metal-ion-mediated base pair for electron transfer in DNA. Chem. Eur. J. 2013, 19, 12547–12552. [Google Scholar] [CrossRef] [PubMed]
- Léon, J.C.; Stegemann, L.; Peterlechner, M.; Litau, S.; Wilde, G.; Strassert, C.A.; Müller, J. Formation of silver nanoclusters from a DNA template containing Ag(I)-mediated base pairs. Bioinorg. Chem. Appl. 2016. [Google Scholar] [CrossRef] [PubMed]
- Taherpour, S.; Golubev, O.; Lönnberg, T. On the feasibility of recognition of nucleic acid sequences by metal-ion-carrying oligonucleotides. Inorg. Chim. Acta 2016. [Google Scholar] [CrossRef]
- Taherpour, S.; Golubev, O.; Lönnberg, T. Metal-ion-mediated base pairing between natural nucleobases and bidentate 3,5-dimethylpyrazolyl-substituted purine ligands. J. Org. Chem. 2014, 79, 8990–8999. [Google Scholar] [CrossRef] [PubMed]
- Taherpour, S.; Lönnberg, H.; Lönnberg, T. 2,6-Bis(functionalized) purines as metal-ion-binding surrogate nucleobases that enhance hybridization with unmodified 2′-O-methyl oligoribonucleotides. Org. Biomol. Chem. 2013, 11, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Taherpour, S.; Lönnberg, H. Metal ion chelates as surrogates of nucleobases for the recognition of nucleic acid sequences: The Pd2+ complex of 2,6-bis(3,5-dimethylpyrazol-1-yl)purine riboside. J. Nucleic Acids 2012. [Google Scholar] [CrossRef] [PubMed]
- Golubev, O.; Lönnberg, T.; Lönnberg, H. Metal-ion-binding analogs of ribonucleosides: Preparation and formation of ternary Pd2+ and Hg2+ complexes with natural pyrimidine nucleosides. Helv. Chim. Acta 2013, 96, 1658–1669. [Google Scholar] [CrossRef]
- Sinha, I.; Hepp, A.; Kösters, J.; Müller, J. Metal complexes of 6-pyrazolylpurine derivatives as models for metal-mediated base pairs. J. Inorg. Biochem. 2015, 153, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Estep, K.G.; Josef, K.A.; Bacon, E.R.; Carabateas, P.M.; Rumney, S., IV; Pilling, G.M.; Krafte, D.S.; Volberg, W.A.; Dillon, K.; Dugrenier, N.; et al. Synthesis and Structure-Activity Relationships of 6-Heterocyclic-Substituted Purines as Inactivation Modifiers of Cardiac Sodium Channels. J. Med. Chem. 1995, 38, 2582–2595. [Google Scholar] [CrossRef] [PubMed]
- Scharf, P.; Jash, B.; Kuriappan, J.A.; Waller, M.P.; Müller, J. Sequence-dependent duplex stabilization upon formation of a metal-mediated base pair. Chem. Eur. J. 2016, 22, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Richters, T.; Krug, O.; Kösters, J.; Hepp, A.; Müller, J. A family of “click” nucleosides for metal-mediated base pairing: Unravelling the principles of highly stabilising metal-mediated base pairs. Chem. Eur. J. 2014, 20, 7811–7818. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, K.; Kösters, J.; Müller, J. 4-(2′-Pyridyl)imidazole as an artificial nucleobase in highly stabilizing Ag(I)-mediated base pairs. J. Biol. Inorg. Chem. 2015, 20, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Litau, S.; Müller, J. Mononuclear 1,3-dideazaadenine-Ag(I)-thyminate base pairs. Z. Anorg. Allg. Chem. 2015, 641, 2169–2173. [Google Scholar] [CrossRef]
- Sinha, I.; Kösters, J.; Hepp, A.; Müller, J. 6-Substituted purines containing thienyl or furyl substituents as artificial nucleobases for metal-mediated base pairing. Dalton Trans. 2013, 42, 16080–16089. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Hepp, A.; Müller, J. Unprecedented dinuclear silver(I)-mediated base pair involving the DNA lesion 1,N6-ethenoadenine. Dalton Trans. 2015, 44, 3540–3543. [Google Scholar] [CrossRef] [PubMed]
- Switzer, C.; Sinha, S.; Kim, P.H.; Heuberger, B.D. A purine-like nickel(II) base pair for DNA. Angew. Chem. Int. Ed. 2005, 44, 1529–1532. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-K.; Switzer, C. Bis(6-carboxypurine)-Cu2+: A possibly primitive metal-mediated nucleobase pair. Org. Lett. 2014, 16, 4059–4061. [Google Scholar] [CrossRef] [PubMed]
- Swasey, S.M.; Espinosa Leal, L.; Lopez-Acevedo, O.; Pavlovich, J.; Gwinn, E.G. Silver(I) as DNA glue: Ag+-mediated guanine pairing revealed by removing Watson–Crick constraints. Sci. Rep. 2015, 5, 10163. [Google Scholar] [CrossRef] [PubMed]
- Richters, T.; Müller, J. A metal-mediated base pair with a [2+1] coordination environment. Eur. J. Inorg. Chem. 2014. [Google Scholar] [CrossRef]
- Sinha, I.; Fonseca Guerra, C.; Müller, J. A highly stabilizing silver(I)-mediated base pair in parallel-stranded DNA. Angew. Chem. Int. Ed. 2015, 54, 3603–3606. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, B.D.; Shin, D.; Switzer, C. Two Watson–Crick-like metallo base-pairs. Org. Lett. 2008, 10, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.; Röhl, I.; Seela, F. Ag+-mediated DNA base pairing: Extraordinarily stable pyrrolo-dC–pyrrolo-dC pairs binding two silver ions. J. Org. Chem. 2013, 78, 9457–9463. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.K.; Guo, X.; Mei, H.; Seela, F. Robust silver-mediated imidazolo-dC base pairs in metal DNA: Dinuclear silver bridges with exceptional stability in double helices with parallel and antiparallel strand orientation. Chem. Commun. 2015, 51, 17301–17304. [Google Scholar] [CrossRef] [PubMed]
- Rolland, V.; Kotera, M.; Lhomme, J. Convenient preparation of 2-deoxy-3,5-di-O-p-toluoyl-α-d-erythro-pentofuranosyl chloride. Synth. Commun. 1997, 27, 3505–3511. [Google Scholar] [CrossRef]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Léon, J.C.; Sinha, I.; Müller, J. RETRACTED: 6-Pyrazolylpurine as an Artificial Nucleobase for Metal-Mediated Base Pairing in DNA Duplexes. Int. J. Mol. Sci. 2016, 17, 554. https://doi.org/10.3390/ijms17040554
Léon JC, Sinha I, Müller J. RETRACTED: 6-Pyrazolylpurine as an Artificial Nucleobase for Metal-Mediated Base Pairing in DNA Duplexes. International Journal of Molecular Sciences. 2016; 17(4):554. https://doi.org/10.3390/ijms17040554
Chicago/Turabian StyleLéon, J. Christian, Indranil Sinha, and Jens Müller. 2016. "RETRACTED: 6-Pyrazolylpurine as an Artificial Nucleobase for Metal-Mediated Base Pairing in DNA Duplexes" International Journal of Molecular Sciences 17, no. 4: 554. https://doi.org/10.3390/ijms17040554
APA StyleLéon, J. C., Sinha, I., & Müller, J. (2016). RETRACTED: 6-Pyrazolylpurine as an Artificial Nucleobase for Metal-Mediated Base Pairing in DNA Duplexes. International Journal of Molecular Sciences, 17(4), 554. https://doi.org/10.3390/ijms17040554






