The Long Intron 1 of Growth Hormone Gene from Reeves’ Turtle (Chinemys reevesii) Correlates with Negatively Regulated GH Expression in Four Cell Lines
Abstract
:1. Introduction
2. Results
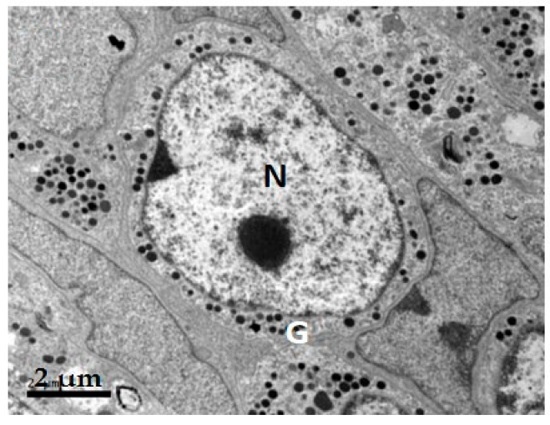
2.1. GH Cells in Reeves’ Turtle Pituitary Glands Contain Few Secretory Granules
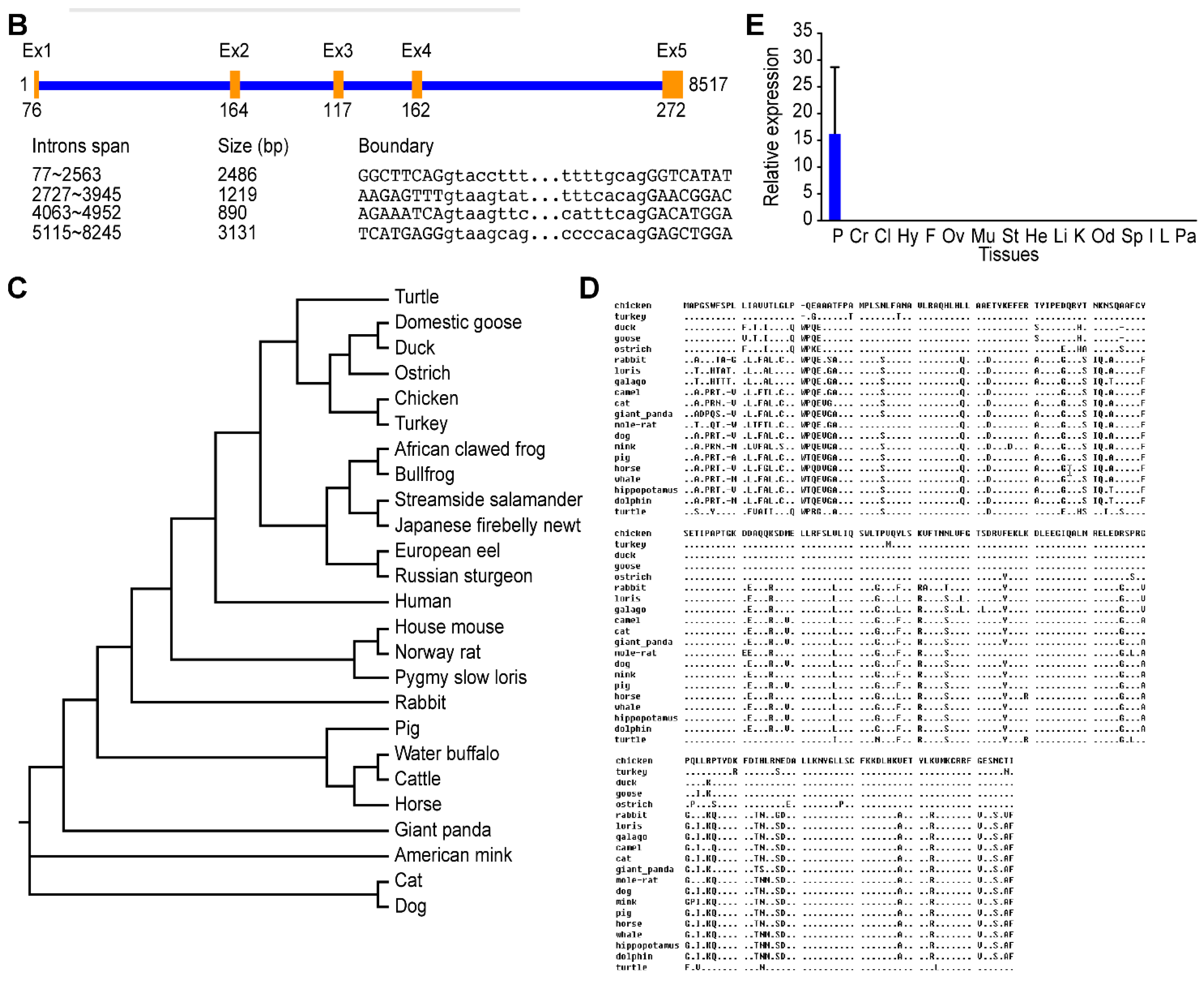
2.2. Sequence and Characteristics of the Reeves’ Turtle GH Gene
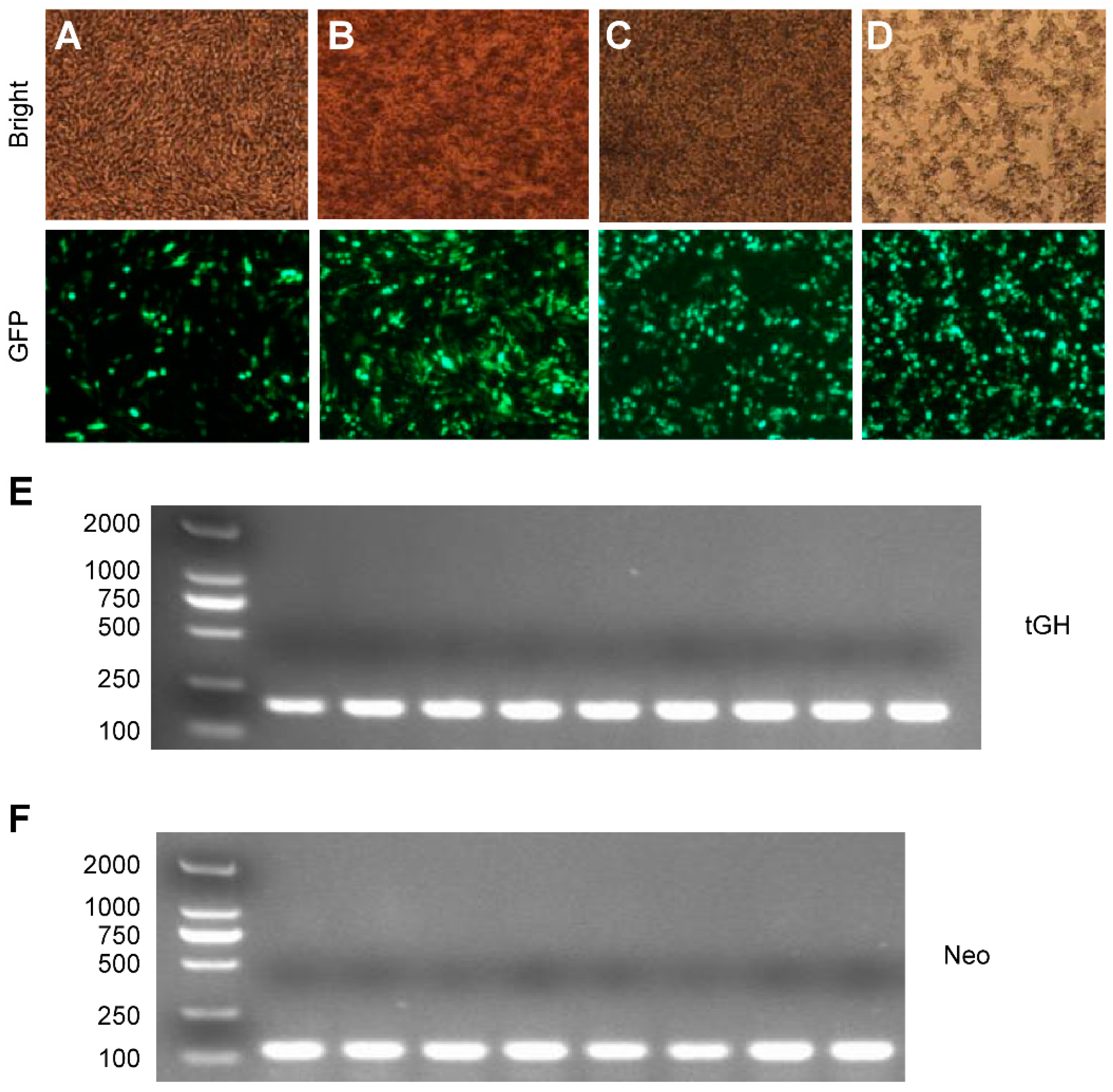
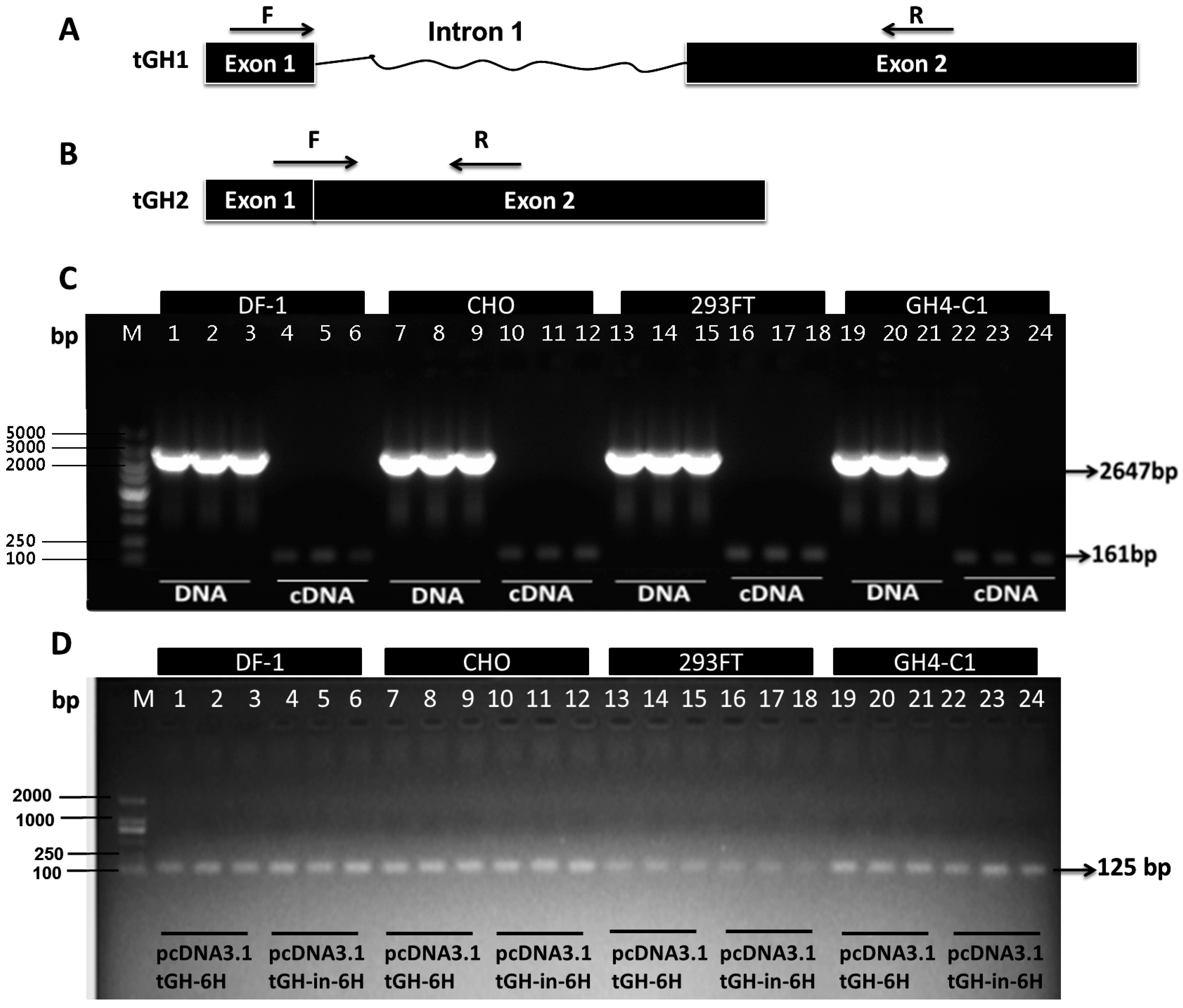
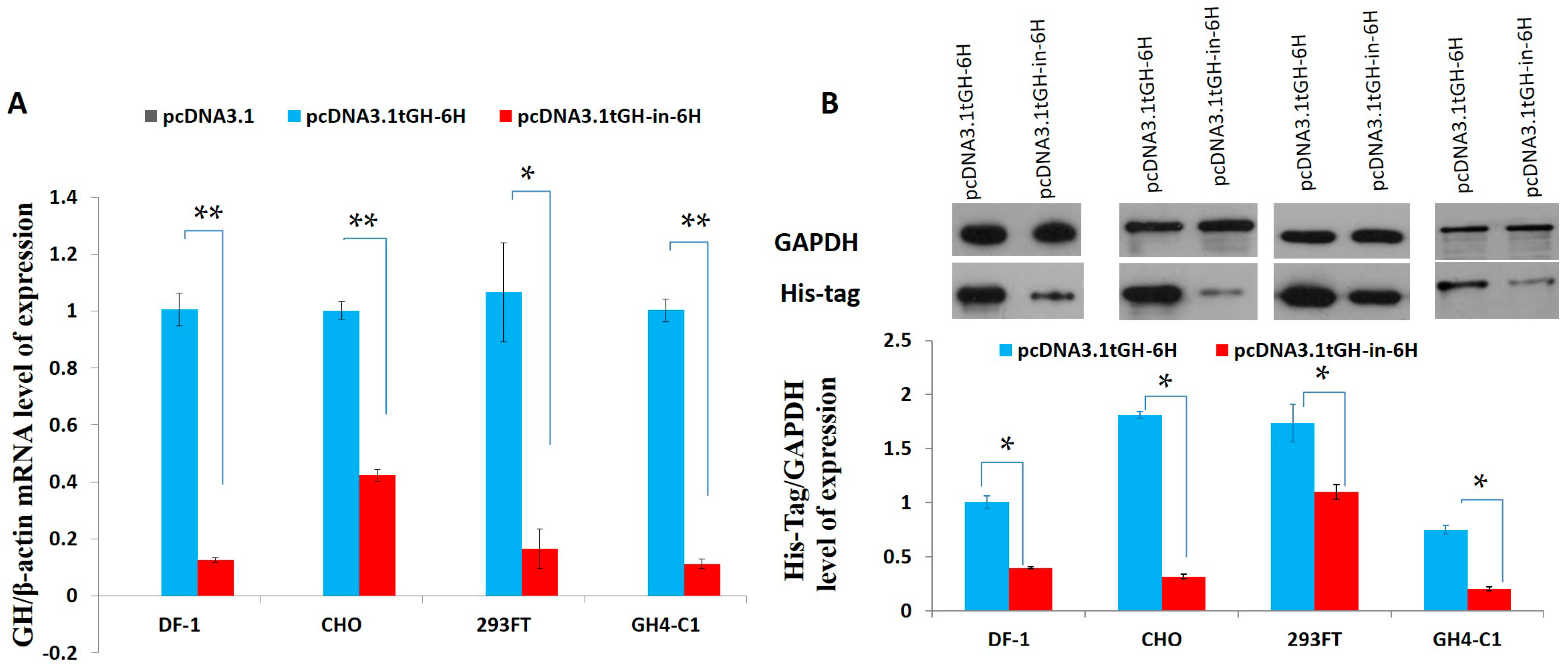
2.3. Effect of Intron 1 on Reeves’ Turtle GH CDS Expression in Four Cell Lines
3. Discussion
4. Experimental Section
4.1. Sample Preparation
4.2. Ultrastructure of Pituitary Gland GH Cells in C. reevesii
4.3. Tissue Collection and Isolation of DNA and RNA
4.4. Cloning, Homology Analysis, and Tissue-Specific Expression Patterns of the Reeves’ Turtle GH Gene
4.5. RT-PCR, RACE, and DNA Sequencing
4.6. Homology Analysis
4.7. Plasmid Construction
4.8. Cell Culture and Transfection
4.9. Real-Time Quantitative PCR Analysis
4.10. Immunoblotting and Immunofluorescence
4.11. Statistical Analysis
4.12. Ethics Standards
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, W.S.; Li, Y. Studies on the micro-structure of pituitary gland and ultrastructure of adenohypophsis in Chinemys reevesii. Acta Hydrobiol. Sin. 2005, 29, 661–666. [Google Scholar]
- Liu, W.S.; Li, Y. Studies on the biochemical indices of blood in Chinemys reevesii. Acta Hydrobiol. Sin. 2006, 30, 499–501. [Google Scholar]
- Yada, T.; Azuma, T.; Takagi, Y. Stimulation of non-specific immune functions in seawater-acclimated rainbow trout, Oncorhynchus mykiss, with reference to the role of growth hormone. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 129, 695–701. [Google Scholar] [CrossRef]
- Yada, T.; Uchida, K.; Kajimura, S. Immunomodulatory effects of prolactin and growth hormone in the tilapia, Oreochromis mossambicus. J. Endocrinol. 2002, 173, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.S.; Ramsey, M.M.; Sonntag, W.E. A critical analysis of the role of growth hormone and IGF-1 in aging and lifespan. Trends Genet. 2002, 18, 295–301. [Google Scholar] [CrossRef]
- Carter, C.S.; Ramsey, M.M.; Ingram, R.L.; Cashion, A.B.; Cefalu, W.T.; Wang, Z.Q.; Sonntag, W.E. Models of growth hormone and IGF-1 deficiency: Applications to studies of aging processes and life-span determination. J. Gerontol. A Biol. Sci. Med. Sci. 2002, 57, B177–B188. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Sackmann-Sala, L.; Kopchick, J.J. Mouse models of growth hormone action and aging: A proteomic perspective. Proteomics 2013, 13, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Charron, M.; Chern, J.-Y.; Wright, W.W. The cathepsin L first intron stimulates gene expression in rat sertoli cells. Biol. Reprod. 2007, 76, 813–824. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wu, Y.; Lan, Z.; Liu, Y.; Zhang, Y. Molecular analysis of the first intron in the bovine myostatin gene. Mol. Biol. Rep. 2011, 38, 4643–4649. [Google Scholar] [CrossRef] [PubMed]
- Ratajewski, M.; de Boussac, H.; Sachrajda, I.; Bacquet, C.; Kovács, T.; Váradi, A.; Pulaski, L.; Arányi, T. ABCC6 expression is regulated by CCAAT/enhancer-binding protein activating a primate-specific sequence located in the first intron of the gene. J. Investig. Dermatol. 2012, 132, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Camp, S.; de Jaco, A.; Zhang, L.; Marquez, M.; de La Torre, B.; Taylor, P. Acetylcholinesterase expression in muscle is specifically controlled by a promoter-selective enhance some in the first intron. J. Neurosci. 2008, 28, 2459–2470. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.Y.Y.; Boudreau-Lariviere, C.; Angus, L.M.; Mankal, F.A.; Jasmin, B.J. An intronic enhancer containing an N-box motif is required for synapse- and tissue-specific expression of the acetylcholinesterase gene in skeletal muscle fibers. Proc. Natl. Acad. Sci. USA 1999, 96, 4627–4632. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Moore, M.L.; McMillin, J.B. A region in the first exon/intron of rat carnitine palmitoyltransferase Iβ is involved in enhancement of basal transcription. Biochem. J. 2002, 362, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Bendixen, M.H.; Nexø, B.A.; Bohr, V.A.; Frederiksen, H.; McGue, M.; Kølvraa, S.; Christensen, K. A polymorphic marker in the first intron of the Werner gene associates with cognitive function in aged Danish twins. Exp. Gerontol. 2004, 39, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; He, A. Ultrastructure of proximal pars distalis of adenohypophysis in Oreochromis nilotica. J. Fish. Sci. China 2002, 9, 207–210. [Google Scholar]
- Zhu, Y.; Wu, X. Ultrastructural comparison of adenohypophysis pars distalis during laying nonlaying and prelaying periods in SI-JI geese. Chin. J. Vet. Sci. 1998, 18, 390–393. [Google Scholar]
- Li, Y.; Zhang, Y.; Kong, X.; Cheng, S.; Huang, R. Ultrastructural study of the adenohypophysis pars distalis of the Beagle dog. Chin. J. Anim. Vet. Sci. 2003, 34, 468–470. [Google Scholar]
- Lin, G.; Fang, Z.; Lin, A. Ultrastructure of the pituitary in Megalobrama terminalis. J. South China Norm. Univ. 2003, 4, 93–99. [Google Scholar]
- Xie, B.W.; Yue, X.J.; Zhang, Y.G.; Lei, A. Studies on the development of the pituitary in Silurus meridionalis. ActaHydrobiol. Sin. 2004, 28, 599–606. [Google Scholar]
- Yang, B.; Greene, M.; Chen, T.T. Early embryonic expression of the growth hormone family protein genes in the developing rainbow trout, Oncorhynchus mykiss. Mol. Reprod. Dev. 1999, 53, 127–134. [Google Scholar] [CrossRef]
- Harvey, S.; Johnson, C.D.M.; Sharma, P.; Sanders, E.J.; Hull, K.L. Growth hormone: A paracrine growth factor in embryonic development? Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998, 119, 305–315. [Google Scholar] [CrossRef]
- Harvey, S.; Johnson, C.D.; Sanders, E.J. Extra-pituitary growth hormone in peripheral tissues of early chick embryos. J. Endocrinol. 2000, 166, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, R.; Kuenzel, W.J.; Buntin, J.D.; Proudman, J.A. Identification of growth-hormone- and prolactin-containing neurons within the avian brain. Cell Tissue Res. 2000, 299, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.E.; Harvey, S. Extrapituitary β TSH and GH in early chick embryos. Mol. Cell. Endocrinol. 2001, 185, 161–171. [Google Scholar] [CrossRef]
- Castillo-Davis, C.I.; Mekhedov, S.L.; Hartl, D.L.; Koonin, E.V.; Kondrashov, F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002, 31, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Benbassat, C.; Shoba, L.N.N.; Newman, M.; Adamo, M.L.; Frank, S.J.; Lowe, W.L., Jr. Growth hormone-mediated regulation of insulin-like growth factor I promoter activity in C6 glioma cells. Endocrinology 1999, 140, 3073–3081. [Google Scholar] [CrossRef] [PubMed]
- Hoeflich, A.; Weber, M.M.; Fisch, T. Insulin-like growth factor binding protein 2 (IGFBP-2) separates hypertrophic and hyperplastic effects of growth hormone (GH)/IGF-I excess on adrenocortical cells in vivo. FASEB J. 2002, 16, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Irimia, M.; Roy, S.W. Origin of spliceosomal introns and alternative splicing. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Niemelä, E.H.; Frilander, M.J. Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol. 2014, 11, 1325–1329. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, S.; Argenton, F.; Vianello, S.; Colombo, L.; Bortolussi, M. Regulatory regions in the promoter and third intron of the growth hormone gene in rainbow trout, Oncorhynchus mykiss walbaum. Gen. Comp. Endocrinol. 1999, 116, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Heuck-Knubel, K.; Proszkowiec-Weglarz, M.; Narayana, J.; Ellestad, L.E.; Prakobsaeng, N.; Porter, T.E. Identification of cis elements necessary for glucocorticoid induction of growth hormone gene expression in chicken embryonic pituitary cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R606–R619. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Pan, J.; Qin, Y.; Lee, D.N.; Bauman, W.A.; Cardozo, C. Identification of functional glucocorticoid response elements in the mouse FoxO1 promoter. Biochem. Biophys. Res. Commun. 2014, 450, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Almuly, R.; Skopal, T.; Funkenstein, B. Regulatory regions in the promoter and first intron of Sparus aurata growth hormone gene: Repression of gene activity by a polymorphic minisatellite. Comp. Biochem. Physiol. D Genom. Proteom. 2008, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Giordano, R.; Bonelli, L.; Marinazzo, E.; Ghigo, E.; Arvat, E. Growth hormone treatment in human ageing: Benefits and risks. Hormones 2008, 7, 133–139. [Google Scholar] [PubMed]
- Bartke, A.; Coschigano, K.; Kopchick, J.; Chandrashekar, V.; Mattison, J.; Kinney, B.; Hauck, S. Genes that prolong life: Relationships of growth hormone and growth to aging and life span. J. Gerontol. A Biol. Sci. Med. Sci. 2001, 56, B340–B349. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A.; Morley, J.E.; Farr, S.A.; Price, T.O.; Ercal, N.; Vidaurre, I.; Schally, A.V. Effects of a growth hormone-releasing hormone antagonist on telomerase activity, oxidative stress, longevity, and aging in mice. Proc. Natl. Acad. Sci. USA 2010, 107, 22272–22277. [Google Scholar] [CrossRef] [PubMed]
- Brown-Borg, H.M.; Borg, K.E.; Meliska, C.J.; Bartke, A. Dwarf mice and the ageing process. Nature 1996, 384, 33. [Google Scholar] [CrossRef] [PubMed]
- Colao, A.; Ferone, D.; Marzullo, P.; Lombardi, G. Systemic complications of acromegaly: Epidemiology, pathogenesis, and management. Endocr. Rev. 2004, 25, 102–152. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Antebi, A.; Bartke, A.; Barzilai, N.; Brown-Borg, H.M.; Caruso, C.; Curiel, T.J.; Cabo, R.; Franceschi, C.; Gems, D.; et al. Interventions to slow aging in humans: Are we ready? Aging Cell 2015, 14, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Maximina, H.Y. Changes in regenerative capacity through lifespan. Int. J. Mol. Sci. 2015, 16, 25392–25432. [Google Scholar]
- Milman, S.; Atzmon, G.; Huffman, D.M.; Wan, J.; Crandall, J.P.; Cohen, P.; Barzilai, N. Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging Cell 2014, 13, 769–771. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiu, H.F.; Hu, W.X.; Li, S.R.; Yu, J.J. Over-expression of The platelet-derived growth factor-D promotes tumor growth and invasion in endometrial cancer. Int. J. Mol. Sci. 2014, 15, 4780–4794. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Zhang, G.R.; Wei, K.J.; Xiong, B.X. Molecular cloning, tissue distribution, and ontogenetic expression of ghrelin and regulation of expression by fasting and refeeding in the grass carp (Ctenopharyngodon idellus). J. Exp. Zool. A Ecol. Genet. Physiol. 2013, 319, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Keane, J.; Tajouri, L.; Gray, B. The effect of growth hormone administration on the regulation of mitochondrial apoptosis in vivo. Int. J. Mol. Sci. 2015, 16, 12753–12772. [Google Scholar] [CrossRef] [PubMed]
- NCBI BLAST program. Available online: http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi (accessed on 15 June 2015).
- ExPaSyProtParam Tool. Available online: http://www.expasy.org/cgi-bin/protscale.pl (accessed on 10 July 2015).
- SignalP 3.0 server. Available online: http://www.cbs.dtu.dk/services/signal P/ (accessed on 10 July 2015).
- TMHMM2.0 software. Available online: http://www.cbs.dtu.dk/services/TMHMM (accessed on 15 July 2015).
- MEGA software package, version 4.0. Available online: http://www.megasoftware.net/ (accessed on 20 July 2015).
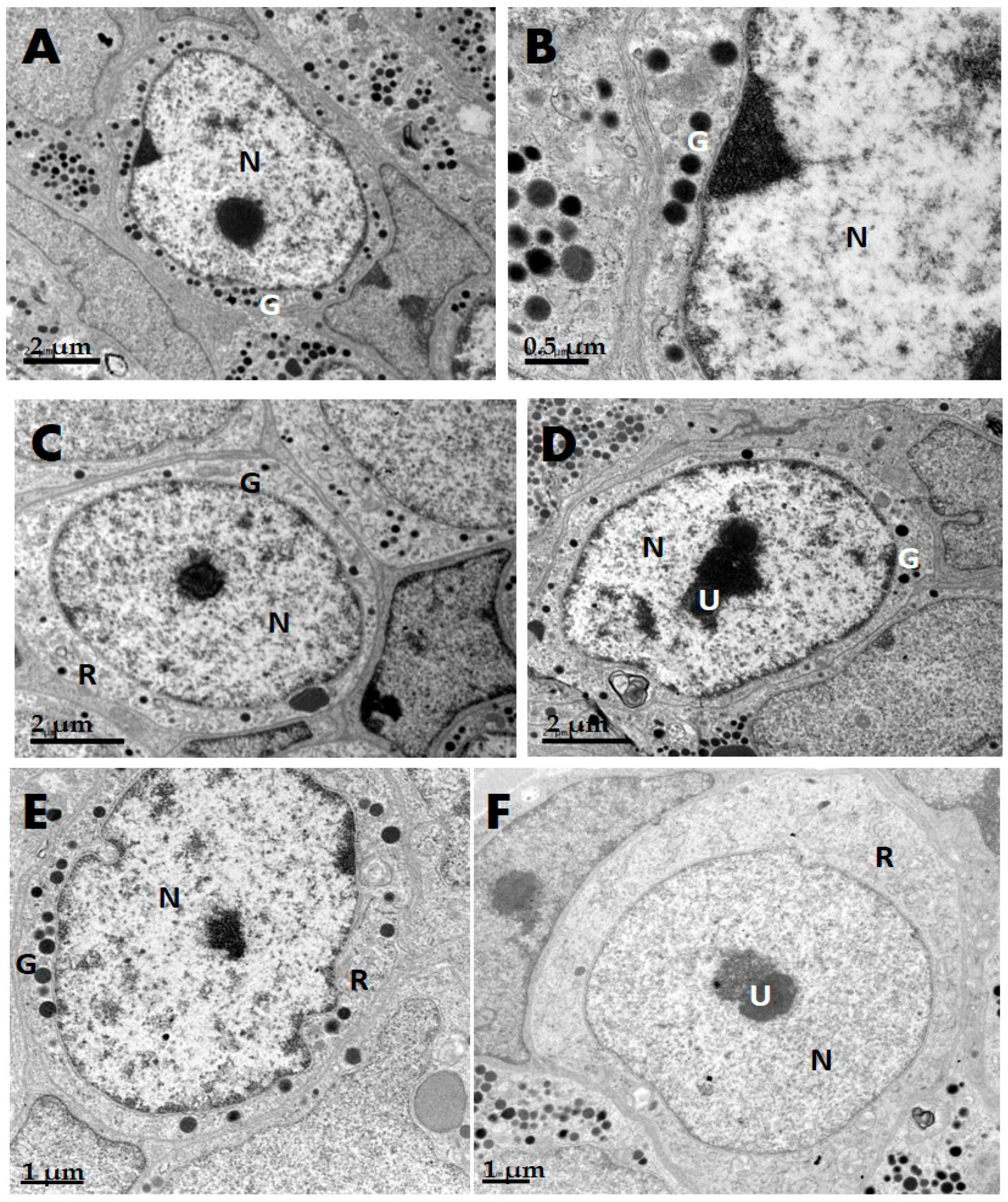

| Species | Cell Shape | Cell Diameter (μm) | Nucleus Shape | Nucleus Diameter (μm) | Secretory Granule, Number, Shape, Size (nm) | Organelle Morphology |
|---|---|---|---|---|---|---|
| Turtle | Round | 8–10 | Round | 5–7 | Few; rare in winter; diameter 250–300 nm | Mitochondria round or oval; a few Golgi apparatuses |
| Oreochromis niloticus [15] | Cuboidal or round | 8–12 | Round | 4–5 | Concentrated in winter, occupation in whole cytoplasm in early spring; diameter 230–350 nm | Mitochondria oval; many Golgi apparatuses; rough endoplasmic reticulum arranged in small fragments |
| Triangle bream [18] | Orbicular-ovate | 11–13 | Round | 4–6 | Many; diameter 220–440 nm | |
| Silurus meridionalis [19] | Irregular | Irregular | concentrated; 150–300 nm | Rough endoplasmic reticulum oval; many Golgi apparatuses | ||
| SI-JI goose [16] | Round or oval | Many; diameter 285 nm | Plenty of mitochondria and rough endoplasmic reticulum | |||
| Beagle dog [17] | Round | Round | Several; diameter 233–465 nm | Rough endoplasmic reticulum, mitochondria, dissociated ribosomes and lysosomes | ||
| Rhesus monkey [17] | Round or oval | Round | Numerous; diameter 200–450 nm |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.-S.; Ma, J.-E.; Li, W.-X.; Zhang, J.-G.; Wang, J.; Nie, Q.-H.; Qiu, F.-F.; Fang, M.-X.; Zeng, F.; Wang, X.; et al. The Long Intron 1 of Growth Hormone Gene from Reeves’ Turtle (Chinemys reevesii) Correlates with Negatively Regulated GH Expression in Four Cell Lines. Int. J. Mol. Sci. 2016, 17, 543. https://doi.org/10.3390/ijms17040543
Liu W-S, Ma J-E, Li W-X, Zhang J-G, Wang J, Nie Q-H, Qiu F-F, Fang M-X, Zeng F, Wang X, et al. The Long Intron 1 of Growth Hormone Gene from Reeves’ Turtle (Chinemys reevesii) Correlates with Negatively Regulated GH Expression in Four Cell Lines. International Journal of Molecular Sciences. 2016; 17(4):543. https://doi.org/10.3390/ijms17040543
Chicago/Turabian StyleLiu, Wen-Sheng, Jing-E Ma, Wei-Xia Li, Jin-Ge Zhang, Juan Wang, Qing-Hua Nie, Feng-Fang Qiu, Mei-Xia Fang, Fang Zeng, Xing Wang, and et al. 2016. "The Long Intron 1 of Growth Hormone Gene from Reeves’ Turtle (Chinemys reevesii) Correlates with Negatively Regulated GH Expression in Four Cell Lines" International Journal of Molecular Sciences 17, no. 4: 543. https://doi.org/10.3390/ijms17040543
APA StyleLiu, W.-S., Ma, J.-E., Li, W.-X., Zhang, J.-G., Wang, J., Nie, Q.-H., Qiu, F.-F., Fang, M.-X., Zeng, F., Wang, X., Lin, X.-R., Zhang, L., Chen, S.-H., & Zhang, X.-Q. (2016). The Long Intron 1 of Growth Hormone Gene from Reeves’ Turtle (Chinemys reevesii) Correlates with Negatively Regulated GH Expression in Four Cell Lines. International Journal of Molecular Sciences, 17(4), 543. https://doi.org/10.3390/ijms17040543