Fine Mapping of Virescent Leaf Gene v-1 in Cucumber (Cucumis sativus L.)
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Characterization of the v-1 Mutant
2.2. High-Resolution Mapping of the v-1 Locus
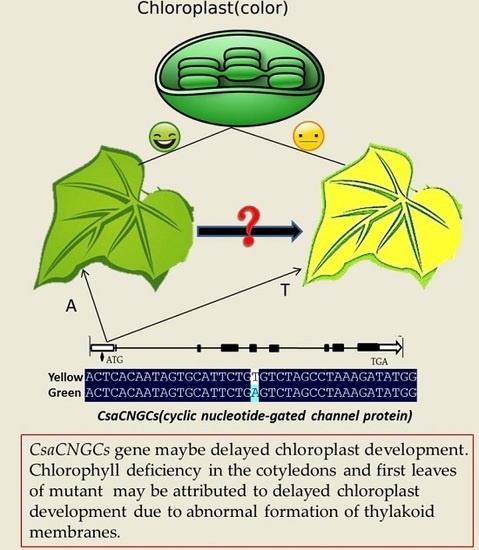
2.3. Candidate Gene Annotation and Sequence Analysis
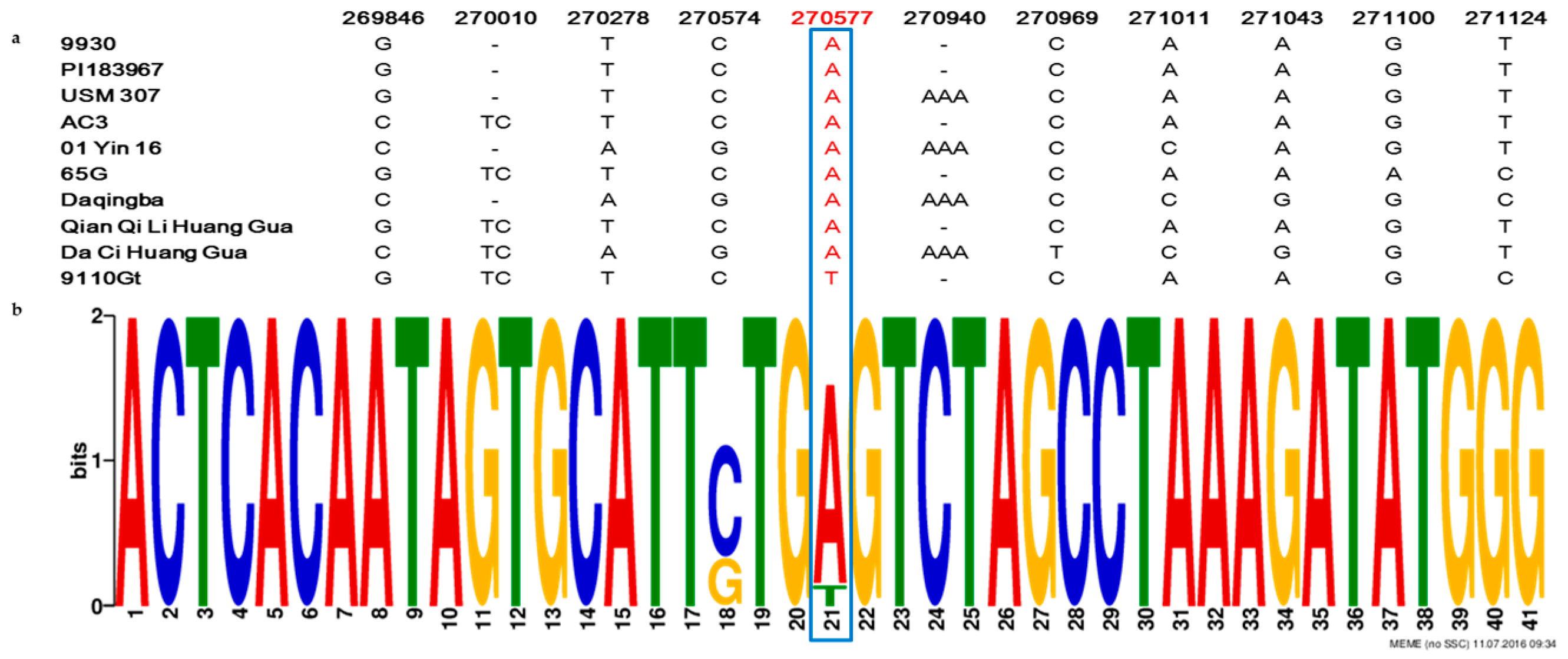
2.4. The Single Nucleotide Mutation in the Promoter Region at the CsaCNGCs Locus Was Unique in Different Cucumber Lines
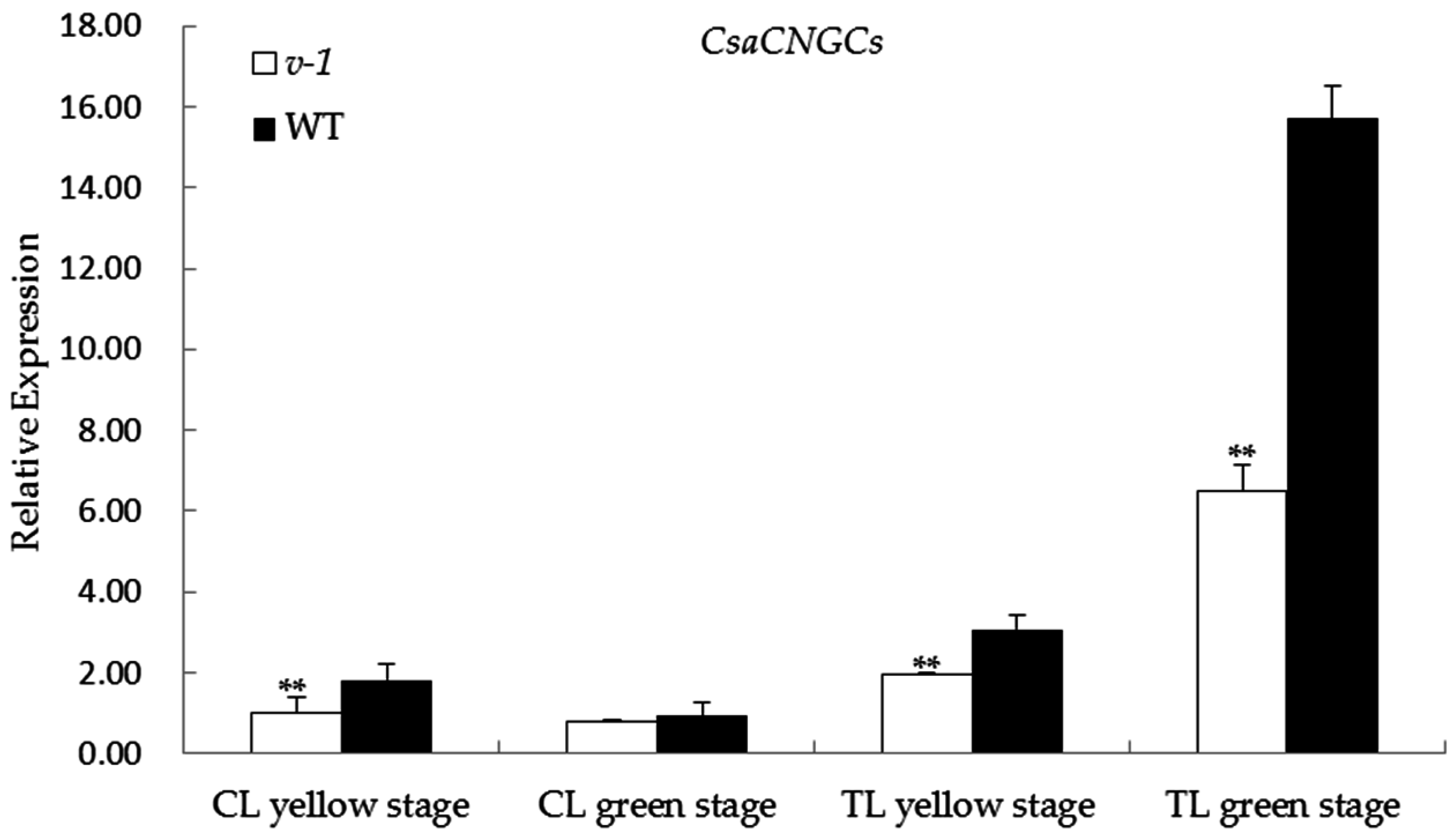
2.5. Expression of CsaCNGCs in 9110Gt Mutant and 9110G WT
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Chlorophyll Contents
4.3. Transmission Electron Microscopy
4.4. Genomic DNA Extraction, PCR, and Electrophoresis
4.5. Development of Molecular Markers for Fine Mapping
4.6. Linkage Mapping and Candidate Gene Annotation
4.7. Real-Time Quantitative PCR of Candidate Genes
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Khan, S.; Wani, R.; Bhat, M.U.D.; Parveen, K. Induced chlorophyll mutations in chickpea (Cicer arietinum L.). Int. J. Agric. Biol. 2005, 5, 764–767. [Google Scholar]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced stay green protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell 2005, 17, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Yuan, S.; Liu, W.J.; Du, J.B.; Tian, W.J.; Luo, M.H.; Lin, H.H. Mutation mechanism of chlorophyll-less barley mutant NYB. Photosynthetica 2008, 46, 73–78. [Google Scholar] [CrossRef]
- Asakura, Y.; Kikuchi, S.; Nakai, M. Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J. 2008, 56, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.T.; Luo, X.D.; Hu, B.L.; Wan, Y.; Xie, J.K. YGL138 (t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 2013, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Q.; Wang, Y.H.; Chai, J.T.; Wang, L.H.; Wang, C.M.; Long, W.; Wang, D.; Wang, Y.; Zheng, M.; Peng, C.; et al. Green-revertible Chlorina 1 (grc1) is required for the biosynthesis of chlorophyll and the early development of chloroplasts in rice. J. Plant Biol. 2013, 56, 326–335. [Google Scholar] [CrossRef]
- Huang, X.Q.; Wang, P.R.; Zhao, H.X.; Deng, X.J. Genetic analysis and molecular mapping of a novel chlorophyll-deficit mutant gene in rice. Rice Sci. 2008, 15, 7–12. [Google Scholar] [CrossRef]
- Reed, S.; Atkinson, T.; Gorecki, C.; Espinosa, K.; Przybylski, S.; Goggi, A.S.; Palmer, R.G.; Sandhu, D. Candidate gene identification for a lethal chlorophyll-deficient mutant in soybean. Agronomy 2014, 4, 462–469. [Google Scholar] [CrossRef]
- Mashkina, E.V.; Usatov, A.V.; Skorina, M.V. Comparative analysis of thermotolerance of sunflower chlorophyll mutants. Russ. J. Genet. 2010, 46, 178–184. [Google Scholar] [CrossRef]
- Hui, Z.; Tian, F.X.; Wang, G.K.; Wang, G.P.; Wang, W. The antioxidative defense system is involved in the delayed senescence in a wheat mutant tasg1. Plant Cell Rep. 2012, 31, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Jia, J.; Xia, C.; Liu, X.; Kong, X. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breed. Sci. 2013, 63, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.J.; Zhang, H.Q.; Wang, Y.; He, F.; Liu, J.L.; Xiao, X.; Wang, G.L. Mapped clone and functional analysis of leaf-color gene ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS ONE 2014, 9, e99564. [Google Scholar] [CrossRef] [PubMed]
- Terry, M.J.; Kendrick, R.E. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green tomato mutants. Plant Physiol. 1999, 119, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.S.; McQuinn, R.P.; Chung, M.Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Nothnagel, T.; Straka, P. Inheritance and mapping of a yellow leaf mutant of carrot (Daucus carota). Plant Breed. 2003, 122, 339–342. [Google Scholar] [CrossRef]
- Budahn, H.; Barański, R.; Grzebelus, D.; Kiełkowska, A.; Straka, P.; Metge, K.; Nothnagel, T. Mapping genes governing flower architecture and pollen development in a double mutant population of carrot. Front. Plant Sci. 2014, 5, 504. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.M.; Zhang, X.; He, B.; Diao, L.P.; Sheng, S.L.; Wang, J.L.; Guo, X.P.; Su, N.; Wang, L.F.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.J.; Zhang, H.Q.; Wang, Y.; Shu, Z.F.; Wang, G.H.; Wang, G.L. Research advances on rice leaf-color mutant genes. Hybrid Rice 2012, 27, 9–14. (In Chinese) [Google Scholar]
- Brusslan, J.A.; Tobin, E.M. Isolation and initial characterization of virescent mutants of Arabidopsis thaliana. Photosynth. Res. 1995, 44, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Maclachlan, S.; Zalik, S. Plastid structure, chlorophyll concentration, and free amino acid composition of a chlorophyll mutant of barley. Can. J. Bot. 1963, 41, 1053–1062. [Google Scholar] [CrossRef]
- Dale, J.E.; Heyes, J.K. A virescent mutant of Phaseolus vulgaris; growth, pigment and plastid characters. New Phytol. 1970, 69, 733–742. [Google Scholar] [CrossRef]
- Benedict, C.R.; McCree, K.J.; Kohel, R.J. High photosynthetic rate of a chlorophyll mutant of cotton. Plant Physiol. 1972, 49, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.R.; Ketring, D.L. Nuclear gene affecting greening in virescent peanut leaves. Plant Physiol. 1972, 49, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Langdale, J.A.; Metzler, M.C.; Nelson, T. The argentia mutation delays normal development of photosynthetic cell-types in Zea mays. Dev. Biol. 1987, 122, 243–255. [Google Scholar] [CrossRef]
- Sugimoto, H.; Kusumi, K.; Noguchi, K.; Yano, M.; Yoshimura, A.; Iba, K. The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 2007, 52, 512–527. [Google Scholar] [CrossRef] [PubMed]
- Archer, E.K.; Bonnett, H.T. Characterization of a virescent chloroplast mutant of tobacco. J. Plant Physiol. 1987, 83, 920–925. [Google Scholar] [CrossRef]
- Pierce, L.K.; Wehner, T.C. Review of genes and linkage groups in cucumber. Hortic. Sci. 1990, 25, 605–615. [Google Scholar]
- Guo, Y.M.; Gu, X.F.; Zhang, C.Z.; Fang, X.J.; Zhang, S.P.; Xu, C.Q. Genetic mechanism of the cucumber leaf mutant. Acta Hortic. Sin. 2003, 30, 409–412. (In Chinese) [Google Scholar]
- Gao, M.L.; Hu, L.L.; Li, Y.H.; Weng, Y.Q. The chlorophyll-deficient golden leaf mutation in cucumber is due to a single nucleotide substitution in CsChlI for magnesium chelatase I subunit. Theor. Appl. Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Zhang, S.P.; Wang, X.W.; Zhang, Z.H.; Li, M.; Mu, S.; Cheng, Z.; Zhang, R.; Huang, S.; Xie, B.; et al. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 2011, 182, 167–176. [Google Scholar] [CrossRef]
- Song, M.; Yang, Z.; Fan, S.; Zhu, H.; Pang, C.; Tian, M.; Yu, S. Cytological and genetic analysis of a virescent mutant in upland cotton (Gossypium hirsutum L.). Euphytica 2012, 187, 235–245. [Google Scholar] [CrossRef]
- Yoo, S.C.; Cho, S.H.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.J.; Iba, K.; Paek, N.C. Rice Virescent-3 and Stripe-1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, K.; Mizutani, A.; Nishimura, M.; Iba, K. A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J. 1997, 12, 1241–1250. [Google Scholar] [CrossRef]
- Kusumi, K.; Sakata, C.; Nakamura, T.; Kawasaki, S.; Yoshimura, A.; Iba, K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 2011, 68, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fei, G.L.; Wu, C.Y.; Wu, F.Q.; Sun, Y.Y.; Chen, M.J.; Ren, Y.L.; Zhou, K.N.; Cheng, Z.J.; Wang, J.L.; et al. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 2013, 162, 1867–1880. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.N.; Ren, Y.L.; Lv, J.; Wang, Y.H.; Liu, F.; Zhou, F.; Zhao, S.; Chen, S.; Peng, C.; Zhang, X.; et al. Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 2013, 237, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Xue, D.X.; Li, X.Y.; Long, Y.M.; Zeng, X.J.; Liu, Y.G. Characterization and molecular mapping of a new virescent mutant in rice. J. Genet. Genom. 2014, 41, 353–356. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Xing, A.Q.; Williams, M.E.; Bourett, T.M.; Hu, W.N.; Hou, Z.L.; Meeley, R.B.; Jaqueth, J.; Dam, T.; Li, B.L. A pair of homoeolog ClpP5 genes underlies a virescent yellow-like mutant and its modifier in maize. Plant J. 2014, 79, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Xu, H.; Zhang, J.Y.; Liang, G.W.; Liu, Y.T.; Guo, A.G. Effect of low temperature on chlorophyll biosynthesis in albinism line of wheat (Triticum aestivum) FA85. Physiol. Plant. 2012, 145, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Fernández, F.; Padmarasu, S.; Surbanovski, N.; Evans, K.M.; Tobutt, K.R.; Sargent, D.J. Characterization of the virescent locus controlling a recessive phenotype in apple rootstocks (Malus pumila Mill.). Mol. Breed. 2014, 33, 373–383. [Google Scholar] [CrossRef]
- Biswal, U.C.; Biswal, B.; Raval, M.K. Chloroplast Biogenesis: From Proplastid to Gerontoplast; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 19–77. [Google Scholar]
- Dare, A.P.; Schaffer, R.J.; Wang, L.K.; Allan, A.C.; Hellens, R.P. Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods 2008, 4, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Koussevitzky, S.; Stanne, T.M.; Peto, C.A.; Giap, T.; Sjogren, L.L.; Zhao, Y.D.; Clarke, A.K.; Chory, J. An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 2007, 63, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.; Moeder, W.; Yoshioka, K. Biological roles of cyclic-nucleotide-gated ion channels in plants: What we know and don’t know about this 20 member ion channel family. Botany 2009, 87, 668–677. [Google Scholar] [CrossRef]
- Schuurink, R.C.; Shartzer, S.F.; Fath, A.; Jones, R.L. Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. USA 1998, 95, 1944–1949. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, Z.; Kakar, K.U.; Saand, M.A.; Shu, Q.Y. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genom. 2014, 15, 853. [Google Scholar] [CrossRef] [PubMed]
- Talke, I.N.; Blaudez, D.; Maathuis, F.J.; Sanders, D. CNGCs: Prime targets of plant cyclic nucleotide signaling. Trends Plant Sci. 2003, 8, 286–293. [Google Scholar] [CrossRef]
- Martinez-Atienza, J.; van Ingelgem, C.; Roef, L.; Maathuis, F.J. Plant cyclic nucleotide signaling. Plant Signal. 2007, 2, 540–543. [Google Scholar] [CrossRef]
- Bowler, C.; Neuhaus, G.; Yamagata, H.; Chua, N.H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 1994, 77, 73–81. [Google Scholar] [CrossRef]
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.F.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Blum, H.; Beier, H.; Gross, H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar] [CrossRef]
- Li, Y.H.; Yang, L.M.; Pathak, M.; Li, D.W.; He, X.M.; Weng, Y.Q. Fine genetic mapping of cp: A recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor. Appl. Genet. 2011, 123, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.M.; Koo, D.H.; Li, Y.H.; Zhang, X.J.; Luan, F.S.; Havey, M.J.; Jiang, J.M.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhang, Z.H.; Liu, J.H.; Staub, J.E.; Han, Y.H.; Cheng, Z.C.; Li, X.F.; Lu, J.Y.; Miao, H.; Knag, H.; et al. Integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 2009, 4, e5795. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.M.; Li, D.W.; Li, Y.H.; Gu, X.F.; Huang, S.W.; Garcia-Mas, J.; Weng, Y. A 1681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol. 2013, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.M.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.W.; Weng, Y. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Bertil, S.; Maskell, D.L. Cushaw: A cuda compatible short read aligner to large genomes based on the burrows-wheeler transform. Bioinformatics 2012, 28, 1830–1837. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Neff, J.D.; Chory, J.; Pepper, A.E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998, 14, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Untergrasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Salamov, A.A.; Solovyev, V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000, 10, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Materials | Chla (mg/g) | Chlb (mg/g) | Caro (mg/g) | Total Chl (mg/g) | Chla/b |
|---|---|---|---|---|---|
| 9110G | 1.38 ± 0.11 ** | 0.40 ± 0.01 ** | 0.39 ± 0.01 * | 1.78 ± 0.01 ** | 3.45 |
| 9110Gt | 0.74 ± 0.02 | 0.26 ± 0.01 | 0.31 ± 0.06 | 1.00 ± 0.21 | 2.85 |
| % of WT | 46.0 | 35.0 | 21.0 | 44.0 | 17.0 |
| Gene | Start | End | Exon | Annotation |
|---|---|---|---|---|
| ORF1 | 263878 | 268887 | 8 | cyclic nucleotide-gated channel |
| ORF2 | 277911 | 280354 | 4 | 3′-5′-exoribonuclease family protein |
| ORF3 | 281291 | 286306 | 9 | receptor-like serine/threonine-protein kinase NCRK |
| ORF4 | 289805 | 293123 | 1 | uncharacterized protein |
| ORF5 | 293407 | 294673 | 1 | uncharacterized protein |
| ORF6 | 299479 | 301054 | 2 | expansin A23 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, H.; Zhang, S.; Wang, M.; Wang, Y.; Weng, Y.; Gu, X. Fine Mapping of Virescent Leaf Gene v-1 in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2016, 17, 1602. https://doi.org/10.3390/ijms17101602
Miao H, Zhang S, Wang M, Wang Y, Weng Y, Gu X. Fine Mapping of Virescent Leaf Gene v-1 in Cucumber (Cucumis sativus L.). International Journal of Molecular Sciences. 2016; 17(10):1602. https://doi.org/10.3390/ijms17101602
Chicago/Turabian StyleMiao, Han, Shengping Zhang, Min Wang, Ye Wang, Yiqun Weng, and Xingfang Gu. 2016. "Fine Mapping of Virescent Leaf Gene v-1 in Cucumber (Cucumis sativus L.)" International Journal of Molecular Sciences 17, no. 10: 1602. https://doi.org/10.3390/ijms17101602
APA StyleMiao, H., Zhang, S., Wang, M., Wang, Y., Weng, Y., & Gu, X. (2016). Fine Mapping of Virescent Leaf Gene v-1 in Cucumber (Cucumis sativus L.). International Journal of Molecular Sciences, 17(10), 1602. https://doi.org/10.3390/ijms17101602