Binding of Sulpiride to Seric Albumins
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fluorescence Quenching
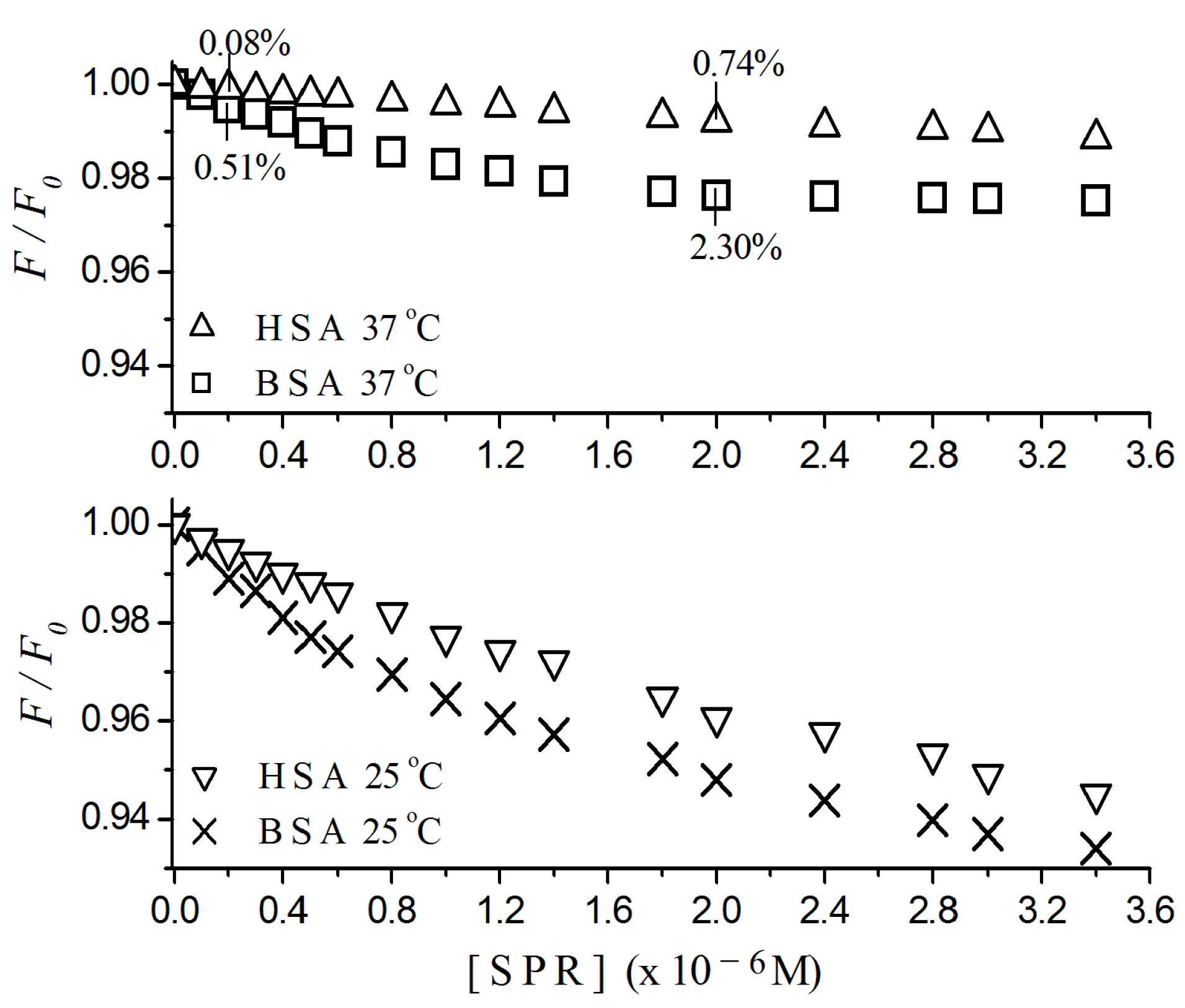
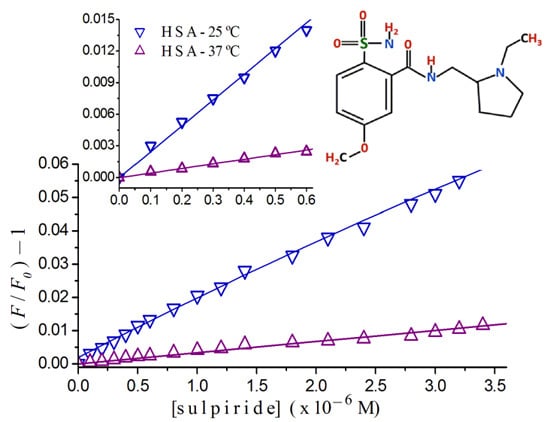
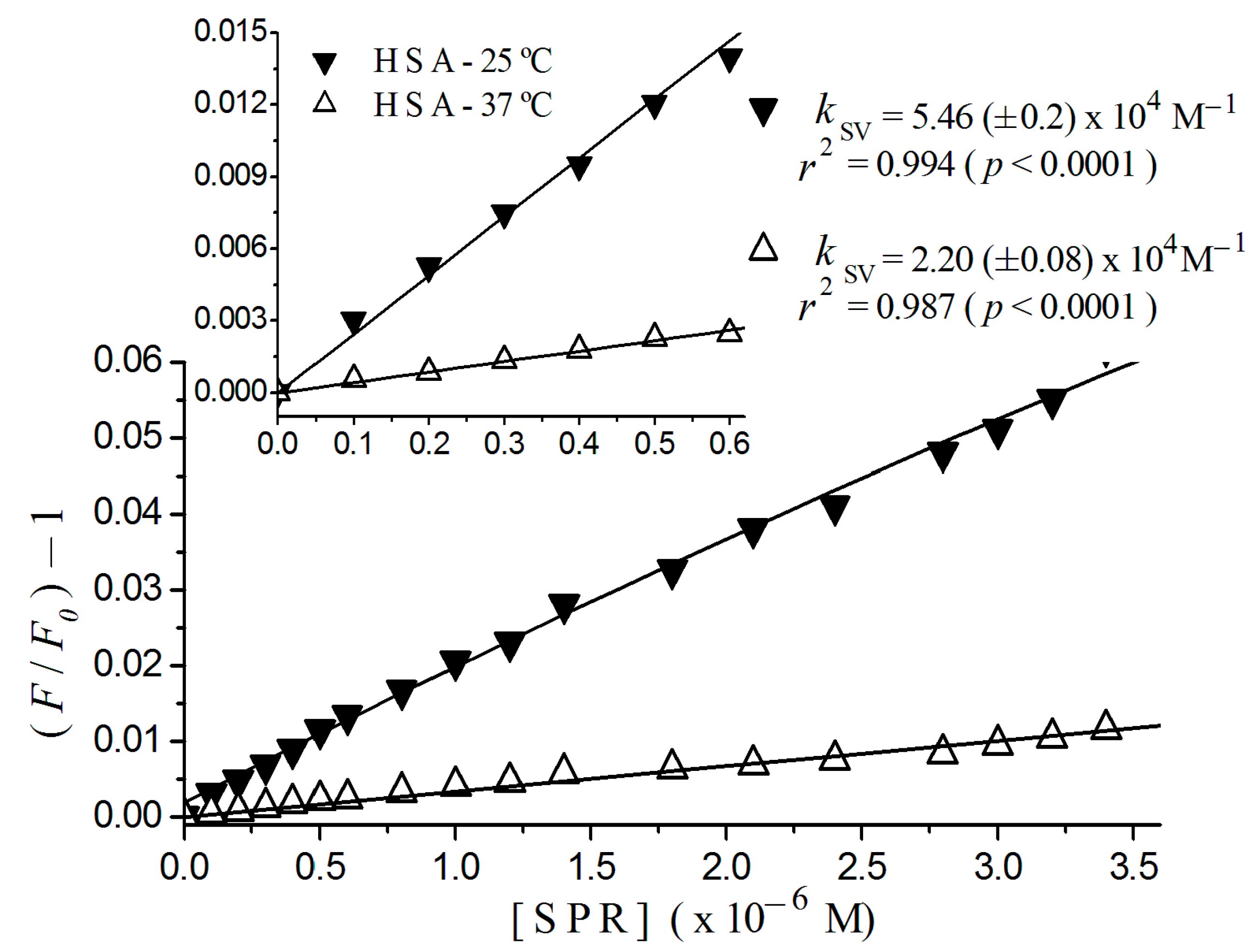
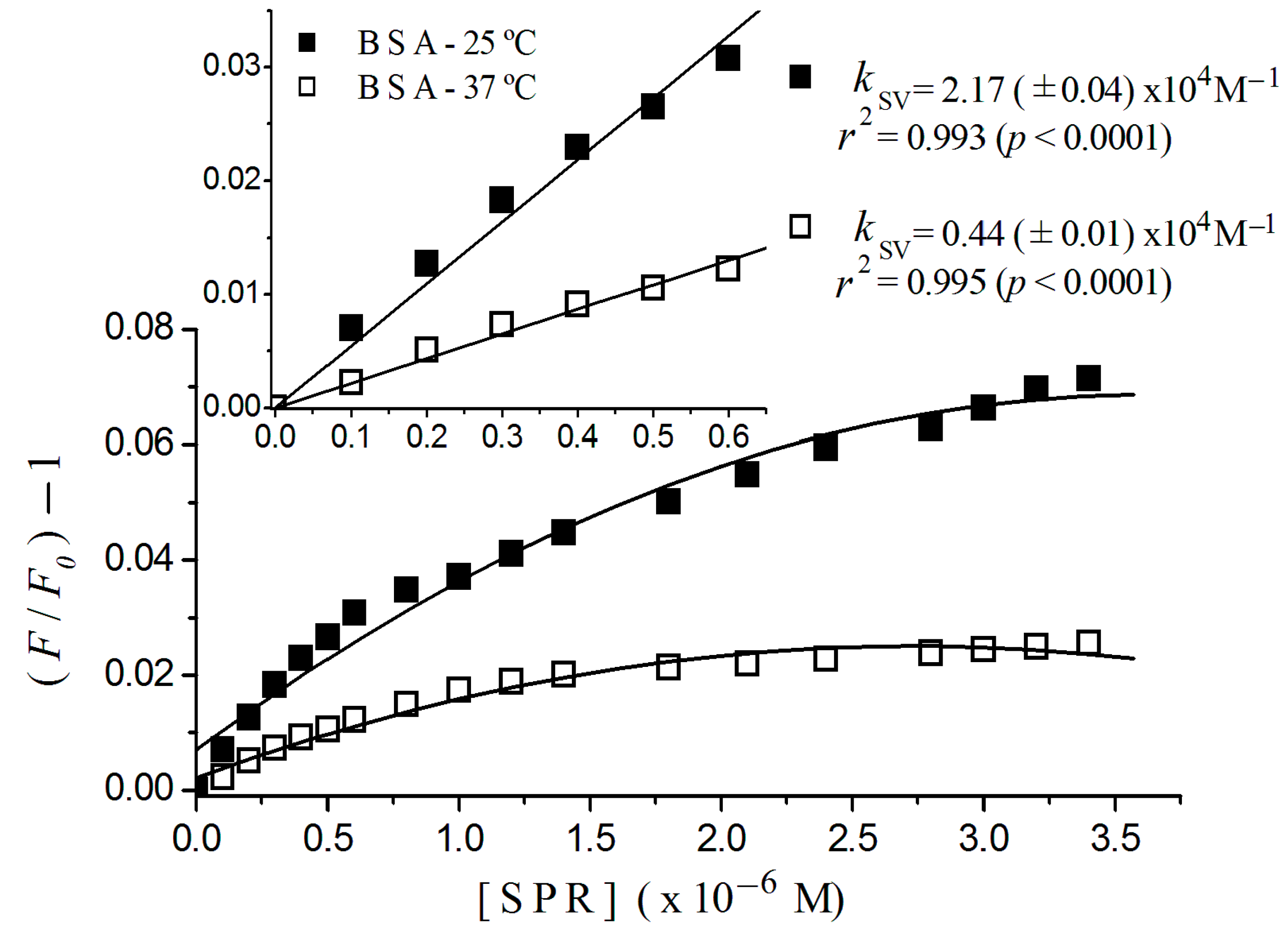
2.2. Stern-Volmer Graphs of Albumins Titrated with Sulpiride
2.3. Fluorescence Quenching of HSA Titrated with Antipsychotics

| Drug | Quenching | Stern-Volmer Constant—KSV (M−1) | |
|---|---|---|---|
| 37 °C | 25 °C | ||
| Sulpiride | Static | 2.20 (±0.08) × 104 | 5.46 (±0.20) × 104 |
| Risperidone * | Static | 1.43 (±0.05) × 105 | 2.56 (±0.09) × 105 |
| Risperidone/Sulpiride | - | 6.5 | 4.6 |
2.4. Binding Constant and the Number of Binding Sites

| Drug | K (M−1) | n |
|---|---|---|
| Sulpiride | 3.890 (±0.003) × 103 | ≈1 |
| Risperidone | 7.460 (±0.010) × 104 | ≈1 |
3. Material and Methods
3.1. Materials
3.2. Binding of Albumin to Sulpiride—Fluorescence Quenching of Albumin by Spectrofluorimetry
3.3. Mathematical Modeling of Albumin Quenching by a Fluorescent Quencher
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Leucht, S.; Corves, C.; Arbter, D.; Engel, R.R.; Li, C.; Davis, J.M. Second-generation versus first-generation antipsychotic drugs for schizophrenia: A meta-analysis. Lancet 2009, 37, 31–41. [Google Scholar] [CrossRef]
- Seeman, P.; van Tol, H.H.M. Dopamine receptor pharmacology. Trends Pharmacol. Sci. 1994, 15, 264–270. [Google Scholar] [CrossRef]
- Bhugra, D. The global prevalence of schizophrenia. PLoS Med. 2005, 2, e151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, B.R.; Torres, K.C.L.; Miranda, D.M.; Motta, B.S.; Scotti-Muzzi, E.; Guimarães, M.M.; Carneiro, D.S.; Rosa, D.V.F.; Souza, R.P.; Reis, H.J.; et al. Lack of effects of typical and atypical antipsychotics in DARPP-32 and NCS-1 levels in PC12 cells overexpressing NCS-1. J. Negat. Results Biomed. 2010, 9, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Peluso, M.J.; Lewis, S.W.; Barnes, T.R.; Jones, P.B. Extrapyramidal motor side-effects of first- and second-generation antipsychotic drugs. Br. J. Psychiatry 2012, 200, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Lerner, V.; Libov, I.; Kotler, M.; Strous, R.D. Combination of “atypical” antipsychotic medication in the management of treatment-resistant schizophrenia and schizoaffective disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 2004, 28, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Mauri, M.C.; Bravin, S.; Bitetto, A.; Rudelli, R.; Invernizzi, G. A risk-benefit assessment of sulpiride in the treatment of schizophrenia. Drug Saf. 1996, 14, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.; Kenyon, P. Atypical antipsychotics, clozapine and sulpiride do not antagonize amphetamine-induced stereotyped locomotion. Psychopharmacology 1994, 114, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Azorin, J.M.; Dassa, D.; Jalfre, M. The atypical neuroleptic concept. Encephale 1992, 18, 453–457. [Google Scholar] [PubMed]
- Jones, P.B.; Barnes, T.R.; Davies, L.; Dunn, G.; Lloyd, H.; Hayhurst, K.P.; Murray, R.M.; Markwick, A.; Lewis, S.W. Randomized controlled trial of the effect on Quality of Life of second- vs. first-generation antipsychotic drugs in schizophrenia: Cost utility of the latest antipsychotic drugs in schizophrenia study (CUtLASS 1). Arch. Gen. Psychiatry 2006, 63, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C.C.; Chia-Hsien, C.; Yang, Y.H.K.; Lin, S.J.; Lin, C.Y. Effectiveness of sulpiride in adult patients with schizophrenia. Schizophr. Bull. 2013, 39, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Leucht, S.; Komossa, K.; Rummel-Kluge, C.; Corves, C.; Hunger, H.; Schmid, F.; Lobos, C.A.; Schwarz, S.; Davis, J.M. A meta-analysis of head-to-head comparisons of second-generation antipsychotics in the treatment of schizophrenia. Am. J. Psychiatry 2009, 166, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Frota, L.H.; Bueno, J.R.; Silva Filho, J.F. Risperidona, amisulpirida, quetiapina e ziprasidona: Comentários finais ao protocolo do Ministério da Saúde para antipsicóticos atípicos de segunda geração. Braz. J. Psychiatry 2001, 50, 337–362. [Google Scholar]
- Fanali, G.; di Mais, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human serum albumin: From bench to bedside. Mol. Asp. Med. 2012, 33, 209–290. [Google Scholar] [CrossRef] [PubMed]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Fanali, G.; Narciso, P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, V.M.S.; Silva, D.; Cruz, F.A.O.; Cortez, C.M. Modeling the process of interaction of risperidone and serum albumin. Environ. Toxicol. Pharmacol. 2012, 33, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer Science: Philadelphia, PA, USA, 2006; pp. 278–327. [Google Scholar]
- Bal, W.; Sokołowska, M.; Kurowska, E.; Faller, P. Binding of transition metal ions to albumin: Sites, affinities and rates. Biochim. Biophys. Acta 2013, 1830, 5444–5455. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.; Cortez, C.M.; Silva, C.M.; Missailidis, S. A fluorescent spectroscopy and modelling analysis of anti-heparanase aptamers–serum protein interactions. J. Photochem. Photobiol. B 2013, 127, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Carter, D.C.; Ho, J.X. Structure of serum albumin. Adv. Protein Chem. 1994, 45, 153–203. [Google Scholar] [PubMed]
- Kragh-Hansen, U. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981, 33, 17–53. [Google Scholar] [PubMed]
- Simard, J.R.; Zunszain, P.A.; Ha, C.E.; Yang, J.S.; Bhagavan, N.V.; Petitpas, I.; Curry, S.; Hamilton, J.A. Locating high-affinity fatty acid-binding sites on albumin by X-ray crystallography and NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 17958–17963. [Google Scholar] [CrossRef] [PubMed]
- Tavirani, M.; Moghaddamnia, S.; Ranjbar, B.; Amani, M.; Marashi, S. Conformational study of human serum albumin in pre-denaturation temperatures by differential scanning calorimetry, circular dichroism and UV spectroscopy. J. Biochem. Mol. Biol. 2006, 39, 530–536. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Wang, Q.; Xue, W.; Yao, X.; Zhang, Y.; Jin, J. Spectroscopic and molecular modeling evidence of clozapine binding to human serum albumin at subdomain IIA. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 79, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Shaikh, S.M.T.; Manjunatha, S.D.; Seetharamappa, J.; Nagaralli, S.J. Spectroscopic studies on the binding of bioactive phenothiazine compounds to human serum albumin. J. Photochem. Photobiol. A Chem. 2007, 189, 121–127. [Google Scholar] [CrossRef]
- Kandagal, P.B.; Kalanur, S.S.; Manjunatha, D.H.; Seetharamappa, J. Mechanism of interaction between human serum albumin and N-alkyl phenothiazines studied using spectroscopic methods. J. Pharm. Biomed. Anal. 2008, 47, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.; Cortez, C.M.; Louro, S.R.W. Chlorpromazine Interactions to Sera Albumins. A study by the quenching of fluorescence. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2004, 60, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Fanali, G.; Pariani, G.; Ascenzi, P.; Fasano, M. Allosteric and binding properties of Asp1–Glu382 truncated recombinant human serum albumin—An optical and NMR spectroscopic investigation. FEBS J. 2009, 276, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, K.; Kume, M.; Yamamoto, M.; Takegami, S.; Kitade, T. 19F NMR spectroscopic study on the binding of triflupromazine to bovine and human serum albumins. J. Pharm. Biomed. Anal. 2004, 36, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Kragh-Hansen, U.; Tanase, S.; Nakajou, K.; Mitarai, M.; Iwao, Y.; Maruyama, T.; Otagiri, M. Conformational stability and warfarin-binding properties of human serum albumin studied by recombinant mutants. Biochem. J. 2001, 357, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Chuang, V.T.; Maruyama, T.; Otagiri, M. Albumin–drug interaction and its clinical implication. Biochim. Biophys. Acta 2013, 1830, 5435–5443. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, Y.; Liang, H. Interactive Association of drugs binding to human serum albumin. Int. J. Mol. Sci. 2014, 15, 3580–3595. [Google Scholar] [CrossRef] [PubMed]
- Denisoff, O.; Molle, L. Binding study of benzamides to serum albumins by fluorescence probe technique. Arzneimittelforschung 1978, 28, 2156–2157. [Google Scholar] [PubMed]
- Bressolle, F.; Bres, J.; Blanchin, M.D.; Gomeni, R. Sulpiride pharmacokinetics in humans after intramuscular administration at three dose levels. J. Pharm. Sci. 1984, 73, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, S.; Eriksson, B.; Oxenstierna, G.; Halldin, C.; Farde, L. Suggested minimal effective dose of risperidone based on PET-measured D2 and 5-HT2A receptor occupancy in schizophrenic patients. Am. J. Psychiatry 1999, 156, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xiang, B.; Wang, Y.; Chen, C.; Dong, Y.; Fang, H.; Wang, M. Spectroscopic investigation on the binding of bioactive pyridazinone derivative to human serumalbumin and molecular modeling. Colloids Surf. B Biointerfaces 2008, 65, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Hongwei, Z.; Min, G.; Zhaoxia, Z.; Wenfeng, W.; Guozhong, W. Spectroscopic studies on the interaction between riboflavin and albumins. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2006, 65, 811–817. [Google Scholar]
- Tokunaga, H.; Kudo, K.; Jitsufuchi, N.; Ohtsuka, Y.; Imamura, T. Sensitive determination of sulpiride in human plasma by high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1997, 691, 203–207. [Google Scholar] [CrossRef]
- Puchalski, M.M.; Morra, M.J.; von Wandruszka, R. Assessment of inner filter effect corrections in fluorimetry. Fresenius J. Anal. Chem. 1991, 340, 341–344. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Silva Fragoso, V.M.; De Morais Coura, C.P.; Hoppe, L.Y.; Soares, M.A.G.; Silva, D.; Cortez, C.M. Binding of Sulpiride to Seric Albumins. Int. J. Mol. Sci. 2016, 17, 59. https://doi.org/10.3390/ijms17010059
Da Silva Fragoso VM, De Morais Coura CP, Hoppe LY, Soares MAG, Silva D, Cortez CM. Binding of Sulpiride to Seric Albumins. International Journal of Molecular Sciences. 2016; 17(1):59. https://doi.org/10.3390/ijms17010059
Chicago/Turabian StyleDa Silva Fragoso, Viviane Muniz, Carla Patrícia De Morais Coura, Luanda Yanaan Hoppe, Marília Amável Gomes Soares, Dilson Silva, and Celia Martins Cortez. 2016. "Binding of Sulpiride to Seric Albumins" International Journal of Molecular Sciences 17, no. 1: 59. https://doi.org/10.3390/ijms17010059