Serum Deprivation-Induced Human GM3 Synthase (hST3Gal V) Gene Expression Is Mediated by Runx2 in Human Osteoblastic MG-63 Cells
Abstract
:1. Introduction
2. Results
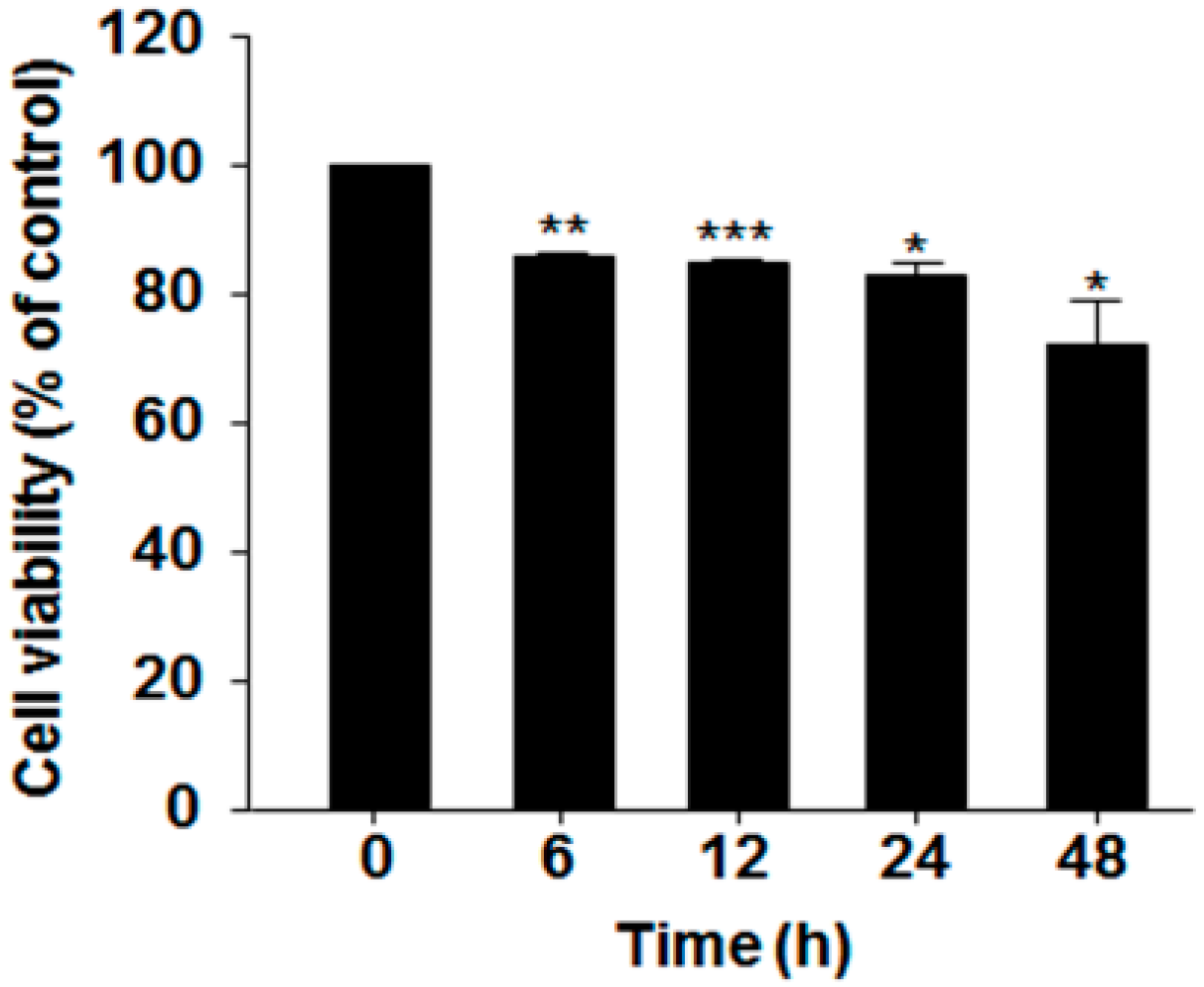
2.1. Effect of Serum Deprivation on Cell Proliferation
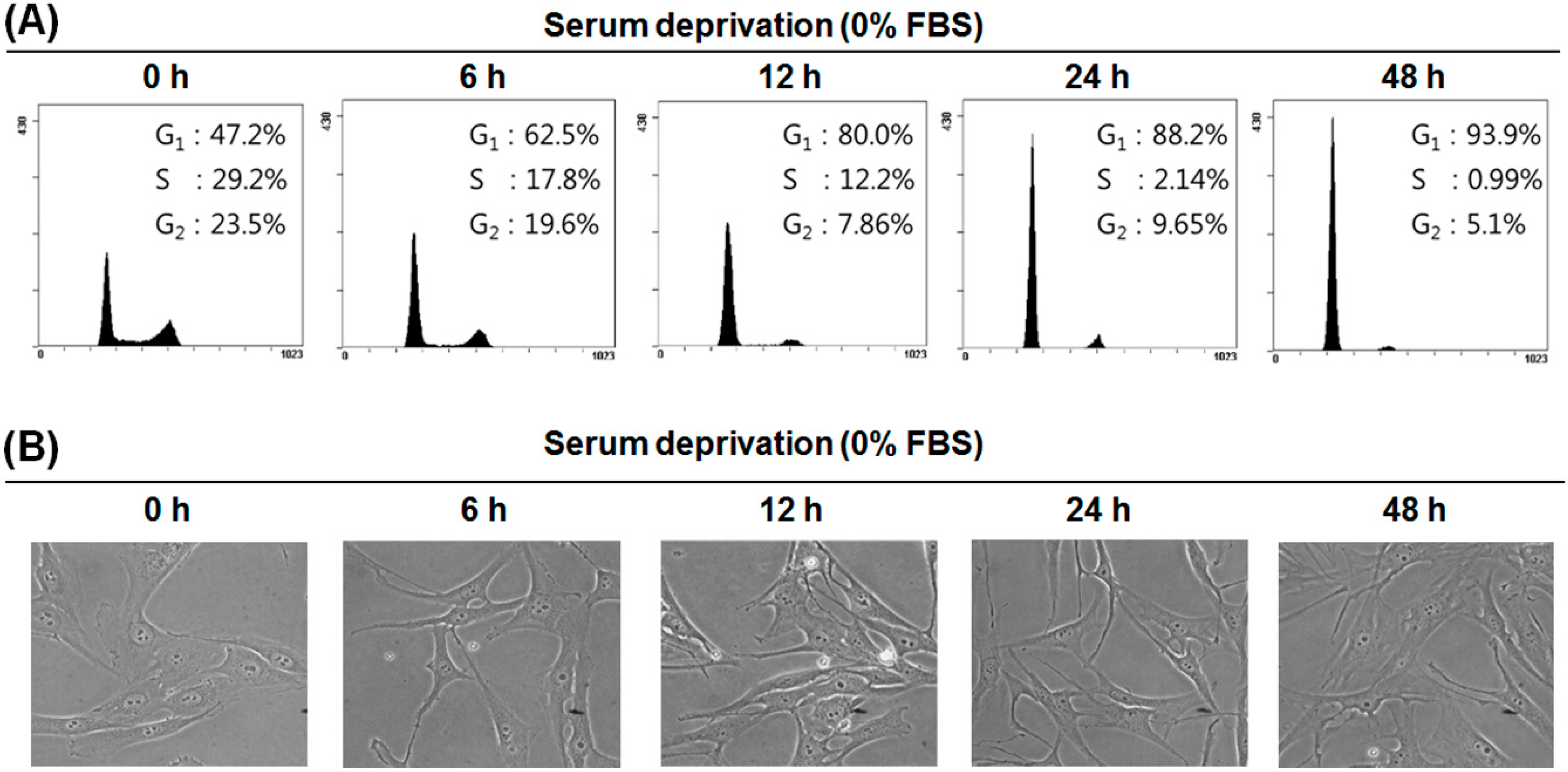
2.2. Serum Deprivation (SD) Induces G1 Arrest of the Cell Cycle in MG-63 Cells
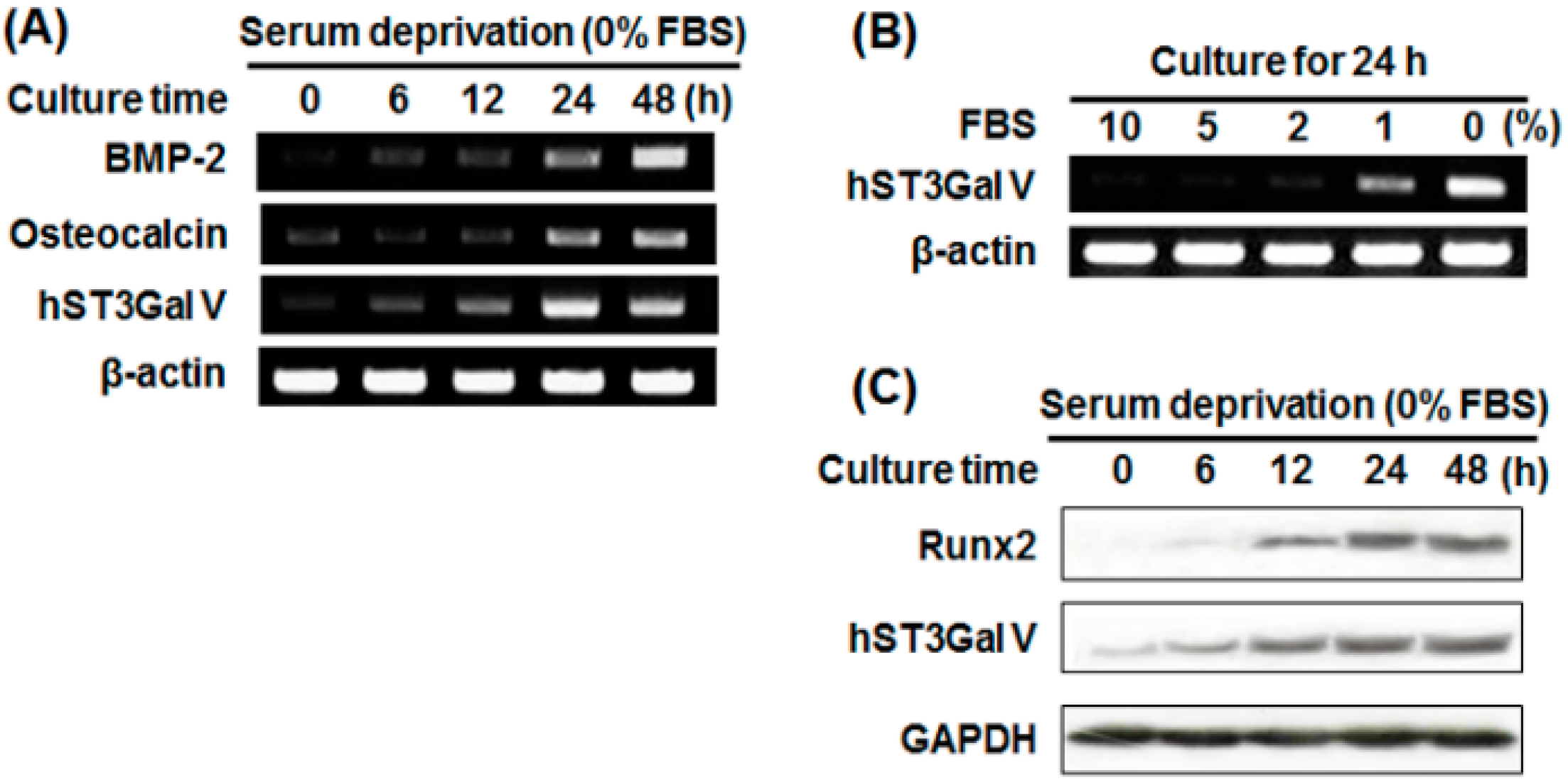
2.3. Effect of SD on Osteoblast-Related Marker Gene Expression in MG-63 Cells
2.4. Effect of SD on hST3Gal V Expression in MG-63 Cells

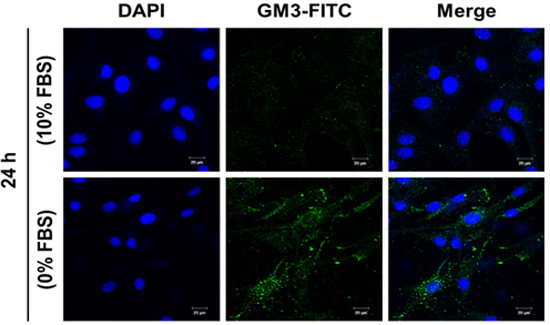
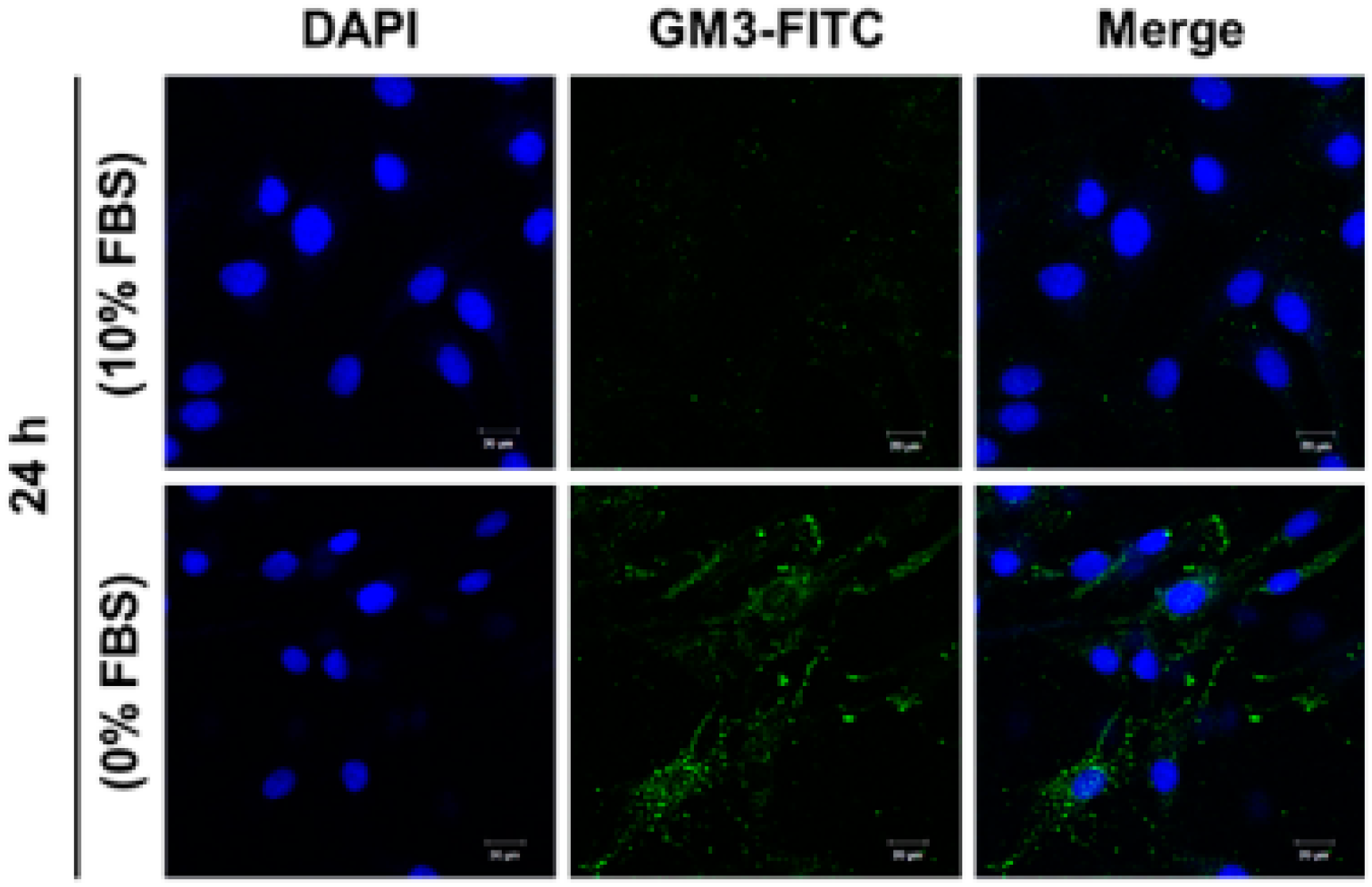
2.5. Effect of SD on Ganglioside GM3 Expression in MG-63 Cells
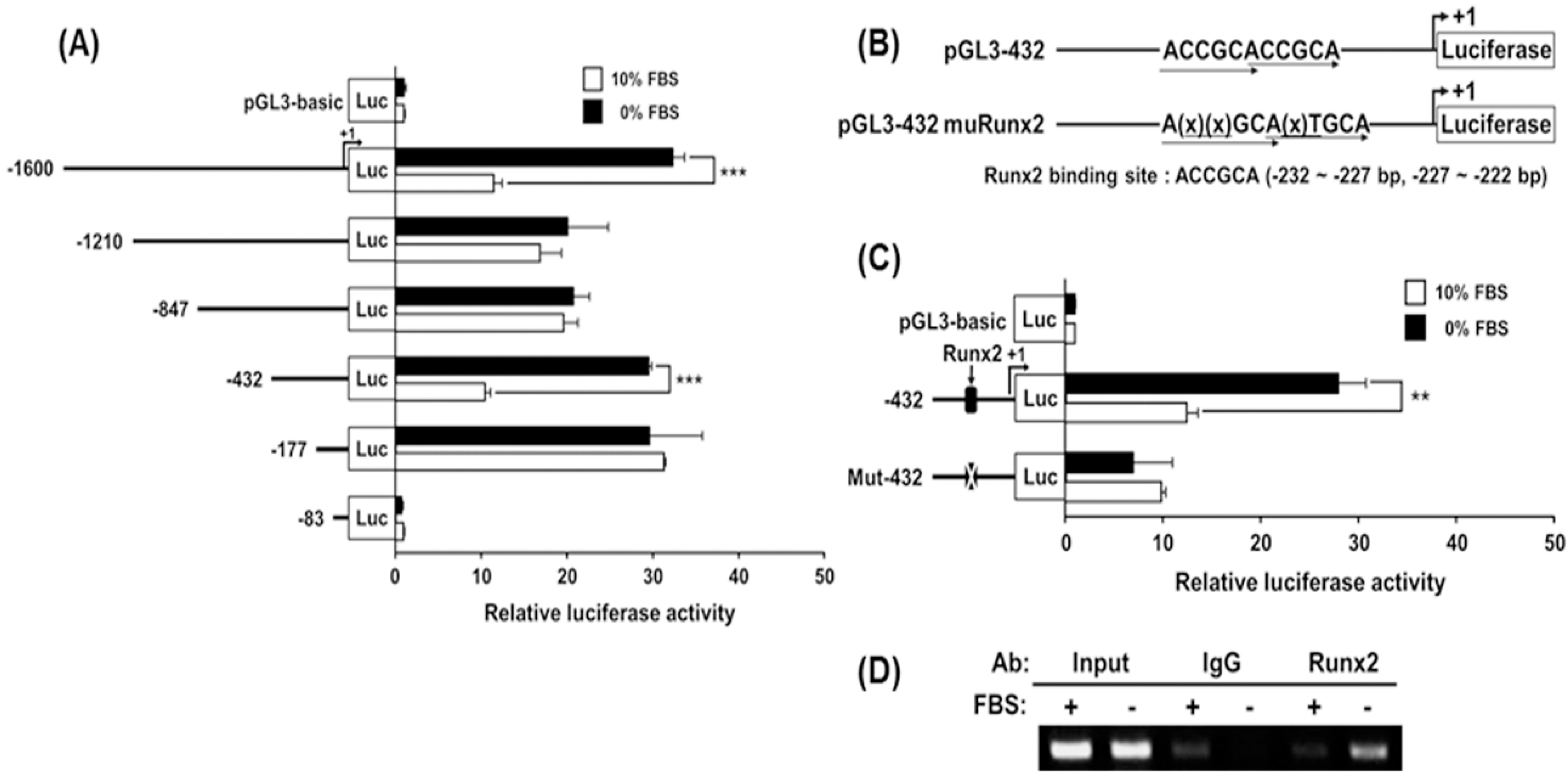
2.6. Analysis of Transcriptional Activity of the hST3Gal V Promoter by SD in MG-63 Cells
2.7. Identification of the SD-Responsive Element in the Functional −432/−178 Region of the hST3Gal V Promoter
3. Discussion
4. Experimental Section
4.1. Cell Cultures
4.2. Cell Viability Assay
4.3. Reverse Transcription-Polymerase Chain Reaction and Quantitative Real-Time PCR
| Primer | Sequence | Strand | Purpose |
|---|---|---|---|
| hST3Gal V | 5′-CCCTGCCATTCTGGGTACGAC-3′ | Sense | RT-PCR |
| hST3Gal V | 5′-CACGATCAATGCCTCCACTGAGA-3′ | Antisense | RT-PCR |
| BMP-2 | 5′-ATGTTCGCCTGAAACAGAGACCCA-3′ | Sense | RT-PCR |
| BMP-2 | 5′-CTTACAGCTGGACTTAAGGCGTTTC-3′ | Antisense | RT-PCR |
| Osteocalcin | 5’-ATGAGAGCCCTCACACTCCTC-3′ | Sense | RT-PCR |
| Osteocalcin | 5′-GCCGTAGAAGCGCCGATAGGC-3′ | Antisense | RT-PCR |
| β-actin | 5′-CAAGAGATGGCCACGGCTGCT-3′ | Sense | RT-PCR |
| β-actin | 5′-TCCTTCTGCATCCTGTCGGCA-3′ | Antisense | RT-PCR |
| hST3Gal V | 5′-GAACTCTTGCCAGAGCACGA-3′ | Sense | qRT-PCR |
| hST3Gal V | 5′-CCCAGTTCTAATCCGTGCAG-3′ | Antisense | qRT-PCR |
| BMP-2 | 5′-GGGTTGGAACTCCAGACTGT-3′ | Sense | qRT-PCR |
| BMP-2 | 5′-GAAGAGTGAGTGGACCCCAG-3′ | Antisense | qRT-PCR |
| Osteocalcin | 5′-GAGGGCAGCGAGGTAGTGAA-3′ | Sense | qRT-PCR |
| Osteocalcin | 5′-GGCTCCCAGCCATTGATACA-3′ | Antisense | qRT-PCR |
| ALP | 5′-CCACGTCTTCACATTTGGTG-3′ | Sense | qRT-PCR |
| ALP | 5′-AGACTGCGCCTAGTAGTTGT-3′ | Antisense | qRT-PCR |
| Runx2 | 5′-CCAGATGGGACTGTGGTTACTG-3′ | Sense | qRT-PCR |
| Runx2 | 5′-CGGAGCTCAGCAGAATAATTTTC-3′ | Antisense | qRT-PCR |
| β-actin | 5′-ACCCACTCCTCCACCTTTGAC-3′ | Sense | qRT-PCR |
| β-actin | 5′-CCTGTTGCTGTAGCCAAATTCG-3′ | Antisense | qRT-PCR |
| muRunx2 | 5′-CTCTGGCAATGCCCAGCATGCAGGGCTGACTGGCCG-3′ | Sense | Mutagenesis |
| muRunx2 | 5′-CGGCCAGTCAGCCCTGCATGCTGGGCATTGCCAGAG-3′ | Antisense | Mutagenesis |
| hST3Gal V | 5′-GCCCCGGGTGCGTCCCTG-3′ | Sense | ChIP |
| hST3Gal V | 5′-AGCGCCGCTCTCGCGCC-3′ | Antisense | ChIP |
4.4. Site-Directed Mutagenesis
4.5. Transfection and Luciferase Assay
4.6. Immunofluorescence
4.7. Western Blot Analysis
4.8. Chromatin Immunoprecipitation Assay
4.9. Flow Cytometry Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cooper, S. Reappraisal of serum starvation, the restriction point, G0 and G1 phase arrest points. FASEB J. 2003, 17, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H.S. Viable offspring deprived from fetal and adult mammalian cells. Nature 1997, 385, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Vackova, I.; Engelova, M.; Marinov, I.; Tomanek, M. Cell cycle synchronization of porcine granulosa cells in G1 stage with mimosine. Anim. Reprod. Sci. 2003, 77, 235–245. [Google Scholar] [CrossRef]
- Kues, W.A.; Anger, M.; Carnwarth, J.W.; Motlik, J.; Nieman, H. Cell cycle synchronization of porcine fibroblast: Effects of serum deprivation and reversible cell cycle inhibitors. Biol. Rep. 2000, 62, 412–419. [Google Scholar] [CrossRef]
- Khammanit, R.; Chantakru, S.; Kitiyanant, Y.; Saikhun, J. Effect of serum starvation and chemical inhibitors on cell cycle synchronization of canine dermal fibroblasts. Theriogenology 2008, 70, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; Hong, S.W.; Lee, S.L.; Kim, T.H.; Park, I.C.; An, S.K.; Lee, W.K.; Lim, J.S.; Kim, K.I.; Yang, Y.; et al. Serum starvation induces G1 arrest through suppression of Skp2-CDK2 and CDK4 in SK-OV-3 cells. Int. J. Oncol. 2008, 32, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Canhoto, A.J.; Chestukhin, A.; Litovchick, L.; DeCaprio, J.A. Phosphorylation of the retinoblastoma-related protein p130 in growth-arrested cells. Oncogene 2000, 19, 5116–5122. [Google Scholar] [CrossRef] [PubMed]
- Rawson, C.L.; Loo, D.T.; Duimstra, J.R.; Hedstrom, O.R.; Schmidt, E.E.; Bames, D.W. Death of serum-free mouse embryo cells caused by epidermal growth factor deprivation. J. Cell Biol. 1991, 113, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Lindenboim, L.; Diamond, R.; Rothenberg, E.; Stein, R. Apoptosis induced by serum deprivation of PC12 cells is not preceded by growth arrest and can occur at each phase of the cell cycle. Cancer Res. 1995, 55, 1242–1247. [Google Scholar] [PubMed]
- Mills, J.C.; Kim, L.H.; Pittman, R.N. Differentiation to an NGF-dependent state and apoptosis following NGF removal both occur asynchronously in cultures of PC12 cells. Exp. Cell Res. 1997, 231, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Maruo, T.; Matsuo, H.; Takekida, S.; Deguchi, J. Serum deprivation-induced apoptosis in cultured porcine granulosa cells is characterized by increased expression of p53 protein Fas antigen and Fas ligand and by decreased expression of PCNA. Endocr. J. 1998, 45, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, M.; Sahin, E.; Johannsen, M.; Somasundaram, R.; Manski, D.; Riecken, E.O.; Schuppan, D. Soluble collagen VI drives serum-starved fibroblasts through S phase and prevents apoptosis via down-regulation of Bax. J. Biol. Chem. 1999, 274, 34361–34368. [Google Scholar] [CrossRef] [PubMed]
- Kues, W.A.; Carnwath, J.W.; Paul, D.; Niemann, H. Cell cycle synchronization of porcine fetal fibroblasts by serum deprivation initiates a nonconventional form of apoptosis. Cloning Stem Cells 2002, 4, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Chauvier, D.; Lecoeur, H.; Langonne, A. Upstream control of apoptosis by caspase-2 in serum-deprived primary neurons. Apoptosis 2005, 10, 1243–1259. [Google Scholar] [CrossRef] [PubMed]
- Mirzamohammadi, S.; Mehrabani, M.; Tekiyehmaroof, N.; Sharifi, A.M. Protective effect of 17β-estradiol on serum deprivation-induced apoptosis and oxidative stress in bone marrow-derived mesenchymal stem cells. Hum. Exp. Toxicol. 2015. [Google Scholar] [CrossRef]
- Carraro-Lacroix, L.R.; Ramirez, M.A.; Zorn, T.M.; Rebouças, N.A.; Malnic, G. Increased NHE1 expression is associated with serum deprivation-induced differentiation in immortalized rat proximal tubule cells. Am. J. Physiol. Ren. Physiol. 2006, 291, F129–F139. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wen, J.K.; Zheng, B.; Cheng, Y.; Zhang, C. Serum deprivation results in redifferentiation of human umbilical vascular smooth muscle cells. Am. J. Physiol. Ren. Physiol. 2006, 291, C50–C58. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Structure, organization, and function of glycosphingolipids in membrane. Curr. Opin. Hematol. 2003, 10, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Sonnino, S.; Prinetti, A. Gangliosides as regulators of cell membrane organization and functions. Adv. Exp. Med. Biol. 2010, 688, 165–184. [Google Scholar] [PubMed]
- Bektas, M.; Spiegel, S. Glycosphingolipids and cell death. Glycoconj. J. 2004, 20, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Jennemann, R.; Grone, H.J. Cell-specific in vivo functions of glycosphingolipids: Lessons from genetic deletions of enzymes involved in glycosphingolipid synthesis. Prog. Lipid Res. 2013, 52, 231–248. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc. Natl. Acad. Sci. USA 2002, 99, 10231–10233. [Google Scholar] [CrossRef] [PubMed]
- Bitton, R.J.; Guthmann, M.D.; Gabri, M.R.; Carnero, A.J.; Alonso, D.F.; Fainboim, L.; Gomez, D.E. Cancer vaccines: An update with special focus on ganglioside antigens. Oncol. Rep. 2002, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Kwak, D.H.; Yu, K.; Kim, S.M.; Lee, D.H.; Kim, S.M.; Jung, J.U.; Seo, J.W.; Kim, N.; Lee, S.; Jung, K.Y.; et al. Dynamic changes of gangliosides expression during the differentiation of embryonic and mesenchymal stem cells into neural cells. Exp. Mol. Med. 2006, 38, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.S.; Ko, K.; Lee, J.W.; Park, S.B.; Byun, S.J.; Jeong, E.J.; Ko, K.; Choo, Y.K. Gangliosides are involved in neural differentiation of human dental pulp-derived stem cells. Biochem. Biophys. Res. Commun. 2009, 387, 266–271. [Google Scholar]
- Kim, S.M.; Jung, J.U.; Ryu, J.S.; Jin, J.W.; Yang, H.J.; Ko, K.; You, H.K.; Jung, K.Y.; Choo, Y.K. Effects of gangliosides on the differentiation of human mesenchymal stem cells into osteoblasts by modulating epidermal growth factor receptors. Biochem. Biophys. Res. Commun. 2008, 371, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.J.; Jung, K.Y.; Kwak, D.H.; Lee, S.H.; Ryu, J.S.; Kim, J.S.; Chang, K.T.; Lee, J.W.; Choo, Y.K. Inhibition of ganglioside GD1a synthesis suppresses the differentiation of human mesenchymal stem cells into osteoblasts. Dev. Growth Differ. 2011, 53, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Bergante, S.; Torretta, E.; Creo, P.; Sessarego, N.; Papini, N.; Piccoli, M.; Fania, C.; Cirillo, F.; Conforti, E.; Ghiroldi, A.; et al. Gangliosides as a potential new class of stem cell markers: The case of GD1a in human bone marrow mesenchymal stem cells. J. Lipid Res. 2014, 55, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.E.; Florine, D.L.; Wille, J.J.; Yun, K. Coupling of growth arrest and differentiation at a distinct state in the G1 phase of the cell cycle: GD. Proc. Natl. Acad. Sci. USA 1982, 79, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.C.; Komori, T.; Ito, Y.; Littman, D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Kim, S.W.; Lee, S.H.; Kim, K.S.; Kim, C.H.; Choo, Y.K.; Lee, Y.C. Isolation and characterization of the promoter region of the human GM3 synthase gene. Biochim. Biophys. Acta 2002, 1578, 84–89. [Google Scholar]
- Choi, H.J.; Chung, T.W.; Kang, N.Y.; Kim, K.S.; Lee, Y.C.; Kim, C.H. Transcriptional regulation of the human GM3 synthase (hST3Gal V) gene during monocytic differentiation of HL-60 cells. FEBS Lett. 2003, 555, 204–208. [Google Scholar] [CrossRef]
- Choi, H.J.; Chung, T.W.; Kang, N.Y.; Kim, K.S.; Lee, Y.C.; Kim, C.H. Involvement of CREB in the transcriptional regulation of the human GM3 synthase (hST3Gal V) gene during megakaryocytoid differentiation of human leukemia K562 cells. Biochem. Biophys. Res. Commun. 2004, 313, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.W.; Choi, H.J.; Lee, Y.C.; Kim, C.H. Molecular mechanism for transcriptional activation of ganglioside GM3 synthase and its function in differentiation of HL-60 cells. Glycobiology 2005, 15, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.Y.; Kang, N.Y.; Dae, H.M.; Kim, K.S.; Kim, C.H.; Do, S.I.; Lee, Y.C. Valproic acid-mediated transcriptional regulation of human GM3 synthase (hST3Gal V) in SK-N-BE(2)-C human neuroblastoma cells. Acta Pharmacol. Sin. 2008, 29, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Song, N.R.; Kim, S.J.; Kwon, H.Y.; Son, S.W.; Kim, K.S.; An, H.B.; Lee, Y.C. Transcriptional activation of human GM3 synthase (hST3Gal V) gene by valproic acid in ARPE-19 human retinal pigment epithelial cells. BMB Rep. 2011, 44, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Chung, T.W.; Choi, H.J.; Kwak, C.H.; Song, K.H.; Suh, S.J.; Kwon, K.M.; Chang, Y.C.; Park, Y.G.; Chang, H.W.; et al. Ganglioside GM3 participates in the TGF-β1-induced epithelial mesenchymal transition of human lens epithelial cells. Biochem. J. 2013, 449, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Galindo, M.; Pratap, J.; Young, D.W.; Hovhannisyan, H.; Im, H.J.; Choi, J.Y.; Lian, J.B.; Stein, J.L.; Stein, G.S.; van Wijnen, A.J. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005, 280, 20274–20285. [Google Scholar] [CrossRef] [PubMed]
- Funato, N.; Ohtani, K.; Ohyama, K.; Kuroda, T.; Nakamura, M. Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Mol. Cell. Biol. 2001, 21, 7416–7428. [Google Scholar] [CrossRef] [PubMed]
- Kilbey, A.; Terry, A.; Jenkins, A.; Borland, G.; Zhang, Q.; Wakelam, M.; Cameron, E.R.; Neil, J.C. Runx regulation of sphingolipid metabolism and survival signaling. Cancer Res. 2010, 70, 5860–5869. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of skeletal development by the Runx family of transcription factors. J. Cell. Biochem. 2005, 95, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Phimphilai, M.; Zhao, Z.; Boules, H.; Roca, H.; Franceschi, R.T. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. 2006, 21, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Deng, C.; Li, Y.P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Kim, K.S.; An, H.K.; Kim, C.H.; Moon, H.I.; Lee, Y.C. Dendropanoxide induces autophagy through ERK1/2 activation in MG-63 human osteosarcoma cells and autophagy inhibition enhances dendropanoxide-induced apoptosis. PLoS ONE 2013, 8, e83611. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, H.-K.; Lee, J.-W.; Kim, K.-S.; Mun, S.-W.; Kim, D.-H.; Kim, H.-J.; Kim, C.-H.; Lee, Y.-C. Serum Deprivation-Induced Human GM3 Synthase (hST3Gal V) Gene Expression Is Mediated by Runx2 in Human Osteoblastic MG-63 Cells. Int. J. Mol. Sci. 2016, 17, 35. https://doi.org/10.3390/ijms17010035
Yoon H-K, Lee J-W, Kim K-S, Mun S-W, Kim D-H, Kim H-J, Kim C-H, Lee Y-C. Serum Deprivation-Induced Human GM3 Synthase (hST3Gal V) Gene Expression Is Mediated by Runx2 in Human Osteoblastic MG-63 Cells. International Journal of Molecular Sciences. 2016; 17(1):35. https://doi.org/10.3390/ijms17010035
Chicago/Turabian StyleYoon, Hyun-Kyoung, Ji-Won Lee, Kyoung-Sook Kim, Seo-Won Mun, Dong-Hyun Kim, Hyun-Jun Kim, Cheorl-Ho Kim, and Young-Choon Lee. 2016. "Serum Deprivation-Induced Human GM3 Synthase (hST3Gal V) Gene Expression Is Mediated by Runx2 in Human Osteoblastic MG-63 Cells" International Journal of Molecular Sciences 17, no. 1: 35. https://doi.org/10.3390/ijms17010035
APA StyleYoon, H.-K., Lee, J.-W., Kim, K.-S., Mun, S.-W., Kim, D.-H., Kim, H.-J., Kim, C.-H., & Lee, Y.-C. (2016). Serum Deprivation-Induced Human GM3 Synthase (hST3Gal V) Gene Expression Is Mediated by Runx2 in Human Osteoblastic MG-63 Cells. International Journal of Molecular Sciences, 17(1), 35. https://doi.org/10.3390/ijms17010035