Isolation of Specific Genomic Regions and Identification of Their Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using the CRISPR System and TAL Proteins
Abstract
:1. Introduction
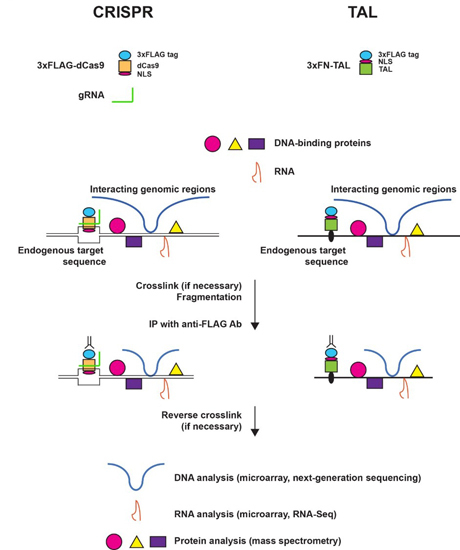
2. The Principle and Applications of Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP)
2.1. Engineered DNA-Binding Molecules
2.2. Scheme of enChIP
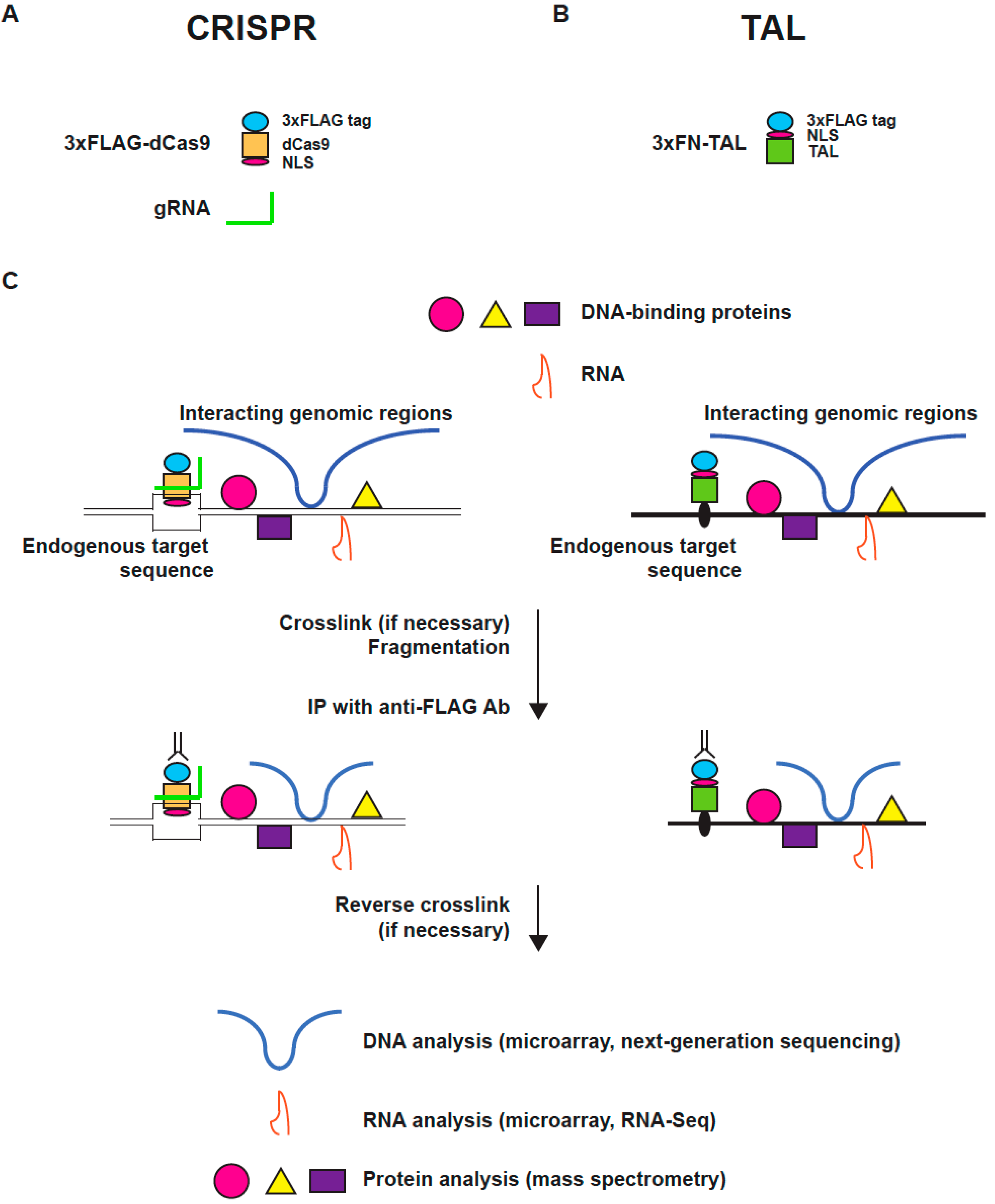
2.3. enChIP-MS
2.4. enChIP-RT-PCR and enChIP-RNA-Seq
2.5. enChIP-Seq
3. Technical Considerations in Performing enChIP
3.1. Design of gRNAs and TAL Proteins
3.2. Transient versus Stable Expression of Engineered DNA-Binding Molecules
| Plasmid | Species | Selection Marker | Addgene ID # | gRNA Plasmid Addgene ID # | Reference |
|---|---|---|---|---|---|
| 3xFLAG-dCas9/ pCMV-7.1 | Mammalian | - | 47948 | 41824 [31] | [18] |
| 3xFLAG-dCas9/ pMXs-puro | Mammalian | Puromycin resistance gene | 51240 | retroviral expression vectors of gRNA * | [19] |
| 3xFLAG-dCas9/ pMXs-neo | Mammalian | Neomycin resistance gene | 51260 | retroviral expression vectors of gRNA * | [19] |
| 3xFLAG-dCas9/ pMXs-IG | Mammalian | GFP | 51258 | retroviral expression vectors of gRNA * | [19] |
| 3xFLAG-dCas9/ pMXs-I2 | Mammalian | hCD2 | 51259 | retroviral expression vectors of gRNA * | [19] |
| 3xFLAG-dCas9/ pTEF1p-CYC1t | Budding yeast | TRP1 | 62190 | 43803 [32] | unpublished |
| 3xFLAG-dCas9/ p-bacteria | Bacteria | Chloramphenicol | 64325 | 44251 [30] | unpublished |
3.3. Controls for enChIP Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dey, B.; Thukral, S.; Krishman, S.; Chakrobarty, M.; Gupta, S.; Manghani, C.; Rani, V. DNA-protein interactions: Methods for detection and analysis. Mol. Cell. Biochem. 2012, 365, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Fujii, H. Insertional chromatin immunoprecipitation: A method for isolating specific genomic regions. J. Biosci. Bioeng. 2009, 108, 446–449. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Fujii, H. Direct idenification of insulator components by insertional chromatin immunoprecipitation. PLoS ONE 2011, 6, e26109. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Fujii, H. Efficient isolation of specific genomic regions by insertional chromatin immunoprecipitation (iChIP) with a second-generation tagged LexA DNA-binding domain. Adv. Biosci. Biotechnol. 2012, 3, 626–629. [Google Scholar] [CrossRef]
- Fujita, T.; Fujii, H. Locus-specific biochemical epigenetics/chromatin biochemistry by insertional chromatin immunoprecipitation. ISRN Biochem. 2013, 2013, 913273. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Fujii, H. Efficient isolation of specific genomic regions retaining molecular interactions by the iChIP system using recombinant exogenous DNA-binding proteins. BMC Mol. Biol. 2014, 15, 26. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Kitaura, F.; Fujii, H. A critical role of the Thy28-MYH9 axis in B cell-specific expression of the Pax5 gene in chicken B cells. PLoS ONE 2015, 10, e0116579. [Google Scholar] [CrossRef] [PubMed]
- McCullagh, E.; Seshan, A.; El-Samad, H.; Madhani, H.D. Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nat. Cell Biol. 2010, 12, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Agelopoulos, M.; McKay, D.J.; Mann, R.S. Developmental regulation of chromain conformation by Hox proteins in Drosophila. Cell Rep. 2012, 1, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Byrum, S.D.; Raman, A.; Taverna, S.D.; Tackett, A. ChAP-MS: A method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012, 2, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Pourfarzad, F.; Aghajanirefah, A.; de Boer, E.; Ten Have, S.; Bryn van Dijk, T.; Kheradmandkia, S.; Stadhouders, R.; Thongjuea, S.; Soler, E.; Gillemans, N.; et al. Locus-specific proteomics by TChP: Targeted chromatin purification. Cell Rep. 2013, 4, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Déjardin, J.; Kingston, R.E. Purification of proteins associated with specific genomic loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Pabo, C.O.; Peisach, E.; Grant, R.A. Design and selection of novel Cys2His2 zinc finger poroteins. Annu. Rev. Biochem. 2001, 70, 313–340. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Voytas, D.F. TAL effectors: Customizable proteins for DNA targeting. Science 2011, 333, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.M.; Jenkins, B.V.; O’Connor-Giles, K.M.; Wildonger, J. A CRISPR view of development. Genes Dev. 2014, 28, 1859–1872. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Fujii, H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 2013, 439, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Fujii, H. Identification of proteins associated with an IFNγ-responsive promoter by a retroviral expression system for enChIP using CRISPR. PLoS ONE 2014, 9, e103084. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Asano, Y.; Ohtsuka, J.; Takada, Y.; Saito, K.; Ohki, R.; Fujii, H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci. Rep. 2013, 3, 3171. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Yuno, M.; Okuzaki, D.; Ohki, R.; Fujii, H. Identification of non-coding RNAs associated with telomeres using a combination of enChIP and RNA sequencing. PLoS ONE 2015, 10, e0123387. [Google Scholar] [CrossRef] [PubMed]
- Nusinzon, I.; Horvath, C.M. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. USA 2003, 100, 14742–14747. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-M.; Paulson, M.; Holko, M.; Rice, C.M.; Williams, B.R.G.; Marié, I.; Levy, D.E. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 9578–9583. [Google Scholar] [CrossRef] [PubMed]
- Chubb, J.R.; Bickmore, W.A. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell 2003, 112, 403–406. [Google Scholar] [CrossRef]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing chromosome conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; de Laat, W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012, 26, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Fujii, H. Identification of proteins interacting with genomic regions of interest in vivo using engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Bio Protoc. 2014, 4, e1124. [Google Scholar]
- Fujita, T.; Fujii, H. Isolation of specific genomic regions and identification of associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Methods Mol. Biol. 2015, 1288, 43–52. [Google Scholar] [PubMed]
- Fujita, T.; Fujii, H. Isolation of specific genomic regions and identification of associated molecules by enChIP. J. Vis. Exp. 2015, in press. [Google Scholar]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Scott, D.A.; Kriz, A.J.; Chiu, A.C.; Hsu, P.D.; Dadon, D.B.; Cheng, A.W.; Trevino, A.E.; Konermann, S.; Chen, S.; et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014, 32, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Cencic, R.; Miura, H.; Malina, A.; Robert, F.; Ethier, S.; Schmeing, T.M.; Dosite, J.; Pelletier, J. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS ONE 2014, 9, e109213. [Google Scholar] [CrossRef] [PubMed]
- O’Green, H.; Henry, I.M.; Bhakta, M.S.; Meckler, J.F.; Segal, D.J. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015, 43, 3389–3404. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujii, H.; Fujita, T. Isolation of Specific Genomic Regions and Identification of Their Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using the CRISPR System and TAL Proteins. Int. J. Mol. Sci. 2015, 16, 21802-21812. https://doi.org/10.3390/ijms160921802
Fujii H, Fujita T. Isolation of Specific Genomic Regions and Identification of Their Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using the CRISPR System and TAL Proteins. International Journal of Molecular Sciences. 2015; 16(9):21802-21812. https://doi.org/10.3390/ijms160921802
Chicago/Turabian StyleFujii, Hodaka, and Toshitsugu Fujita. 2015. "Isolation of Specific Genomic Regions and Identification of Their Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using the CRISPR System and TAL Proteins" International Journal of Molecular Sciences 16, no. 9: 21802-21812. https://doi.org/10.3390/ijms160921802
APA StyleFujii, H., & Fujita, T. (2015). Isolation of Specific Genomic Regions and Identification of Their Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (enChIP) Using the CRISPR System and TAL Proteins. International Journal of Molecular Sciences, 16(9), 21802-21812. https://doi.org/10.3390/ijms160921802