Comparison of Soybean Transformation Efficiency and Plant Factors Affecting Transformation during the Agrobacterium Infection Process
Abstract
:1. Introduction
2. Results
2.1. Transformation Efficiencies of Various Soybean Cultivars
| Cultivar | No. of Explants Infected | No. of Explants with above Three Shoots | Resistant Shoot Induction Rate (%) | No. of Positive T0 Plants | Transformation Efficiency of T0 Plants (%) |
|---|---|---|---|---|---|
| Liaodou16 | 348 | 11 | 3.2 | 0 | 0 |
| General | 481 | 46 | 9.6 | 1 | 0.21 |
| Kottman | 432 | 44 | 10.2 | 3 | 0.69 |
| Dennison | 348 | 67 | 19.3 | 3 | 0.86 |
| Shennong 12 | 504 | 185 | 36.7 | 8 | 1.59 |
| Liaodou 14 | 384 | 244 | 63.5 | 7 | 1.82 |
| Liaodou 10 | 576 | 382 | 66.3 | 13 | 2.26 |
| Bert | 624 | 477 | 76.4 | 32 | 5.13 |
| Shennong 9 | 564 | 436 | 77.3 | 30 | 5.32 |
| Williams 82 | 492 | 397 | 80.7 | 33 | 6.71 |
2.2. Susceptibility of Different Soybean Genotypes to Agrobacterium Infection
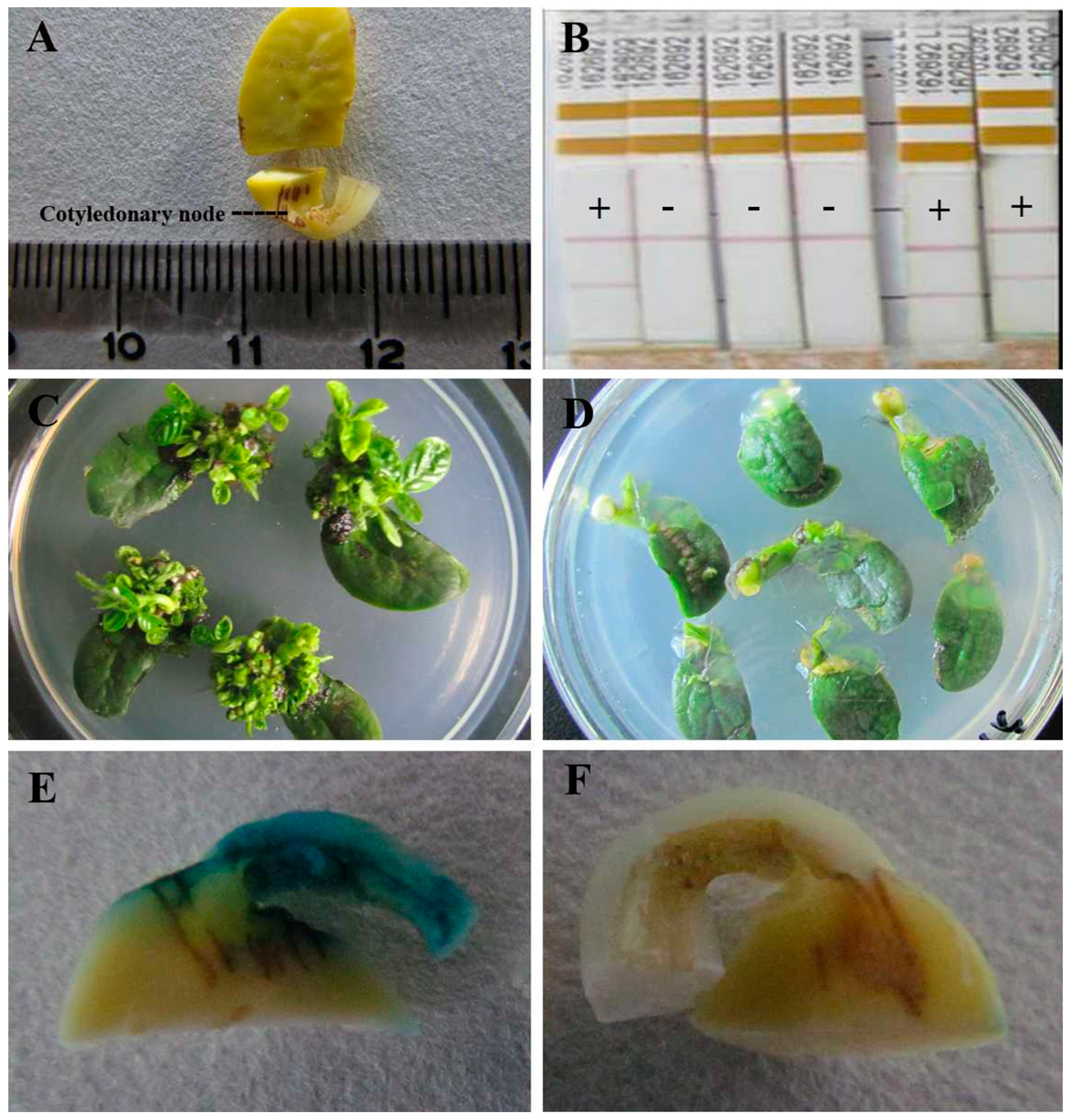
| Genotype | No. of Cotyledonary Nodes for GUS Staining | No. of GUS+ Cotyledonary Nodes | GUS+ Rate (%) |
|---|---|---|---|
| General | 98 | 32 | 32.7 |
| Liaodou 16 | 84 | 16 | 19.1 |
| Kottman | 100 | 30 | 30.0 |
| Williams 82 | 108 | 86 | 85.7 |
| Shennong 9 | 98 | 84 | 79.6 |
| Bert | 104 | 80 | 76.9 |

2.3. Germination and Growth Related Factors Affecting Transformation

2.4. Cell Division Related Factors Affecting Transformation



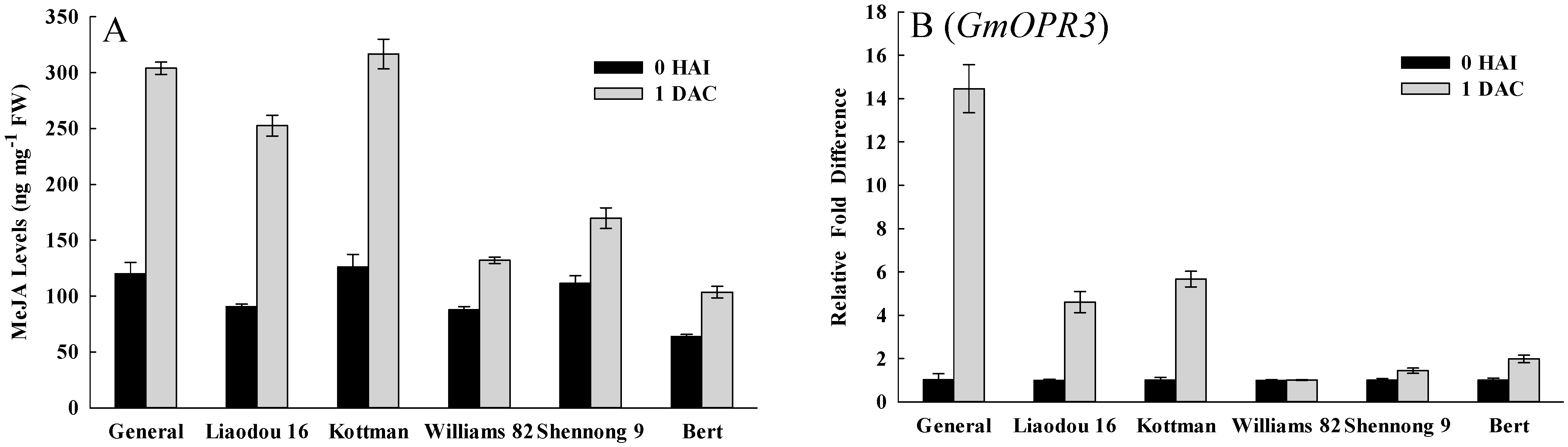
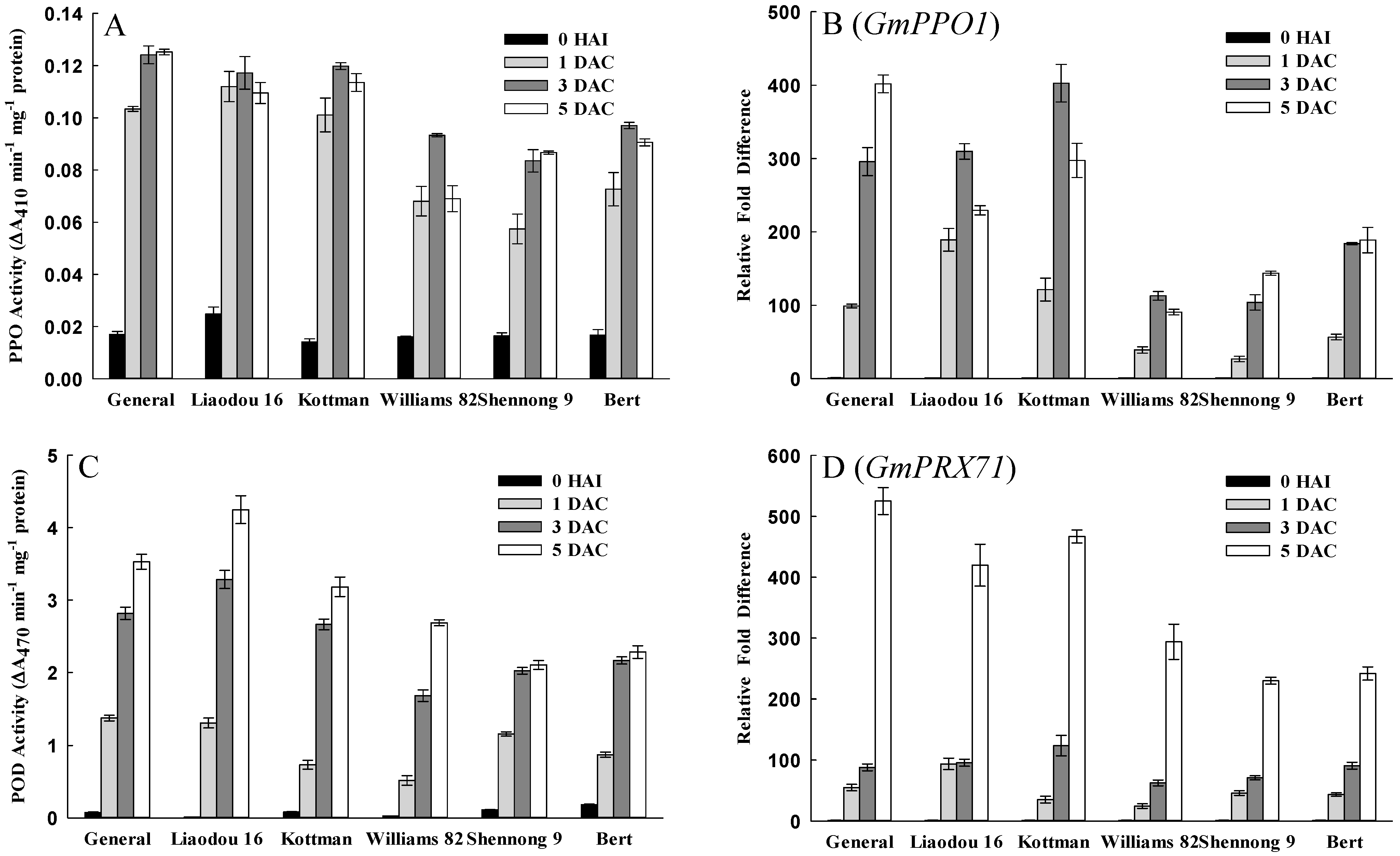
2.5. Defense Response Related Factors Affecting Transformation
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Agrobacterium Preparation
4.3. Infection and Co-Cultivation of Explant
4.4. Transgenetic Shoots Induction and Plant Regeneration
4.5. Detection of T0 Plants
4.6. β-Glucuronidase (GUS) Staining of Cotyledonary Node
4.7. BAR Gene Analysis during Co-Cultivation Period
4.8. Measurement of Gibberellin (GA), Abscisic Acid (ABA), Zeatin Riboside (ZR) and Methyl Jasmonate (MeJA) Content
4.9. Assay of Polyphenol Oxidase (PPO) and Peroxidase (POD) Activity
4.10. Expression of Selected Genes
4.11. Measurement of DNA Quantity
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- James, C. Global status of commercialized biotech/GM crops: 2012. In The International Service for the Acquisition of Agri-Biotech Applications Brief; The International Service for the Acquisition of Agri-biotech Applications: Ithaca, NY, USA, 2012; No. 44. [Google Scholar]
- Miklos, J.A.; Alibhai, M.F.; Bledig, S.A.; Connor-Ward, D.C.; Gao, A.G.; Holmes, B.A.; Kolacz, K.H.; Kabuye, V.T.; MacRae, T.C.; Paradise, M.S.; et al. Characterization of soybean exhibiting high expression of a synthetic Bacillus thuringiensis cry1A transgene that confers a high degree of resistance to lepidopteran pests. Crop Sci. Soc. Am. 2007, 47, 148–157. [Google Scholar]
- Yu, H.; Li, Y.; Li, X.; Romeis, J.; Wu, K. Expression of Cry1Ac in transgenic Bt soybean lines and their efficiency in controlling lepidopteran pests. Pest Manag. Sci. 2013, 69, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Meurer, C.A.; Dinkins, R.D.; Collins, G.B. Factors affecting soybean cotyledonary node transformation. Plant Cell Rep. 1998, 18, 180–186. [Google Scholar] [CrossRef]
- Song, Z.Y.; Tian, J.L.; Fu, W.Z.; Li, L.; Lu, L.H.; Zhou, L.; Shan, Z.H.; Tang, G.X.; Shou, H.X. Screening Chinese soybean genotypes for Agrobacterium-mediated genetic transformation suitability. J. Zhejiang Univ. Sci. B 2013, 14, 289–298. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Travella, S.; Ross, S.M.; Harden, J.; Everett, C.; Snape, J.W.; Harwood, W.A. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep. 2005, 23, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Tzfira, T.; Citovsky, V. From host recognition to T-DNA integeration: The function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol. Plant Pathol. 2000, 1, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Zia, M.; Rizvi, Z.F.; Rehman, R.U.; Chaudhary, M.F. Agrobacterium mediated transformation of soybean (Glycine max L.): Some conditions standardization. Pak. J. Bot. 2010, 42, 2269–2279. [Google Scholar]
- Hobbs, S.L.A.; Jackson, J.A.; Mahon, J.D. Specificity of strain and genotype in the susceptibility of pea to Agrobacterium tumefaciens. Plant Cell Rep. 1989, 8, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, K.; Kaku, H.; Tanaka, H.; Koga-Ban, Y.; Fukuda, M. Enhanced soybean infection by the legume “super-virulent” Agrobacterium tumefaciens strain KAT23. Biosci. Biotechnol. Biochem. 2008, 72, 1809–1816. [Google Scholar] [CrossRef] [PubMed]
- Gelvin, S.B. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Atmakuri, K.; Krishnamoorthy, V.; Jakubowski, S.; Cascales, E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005, 59, 451–485. [Google Scholar] [CrossRef] [PubMed]
- McCullen, C.A.; Binns, A.N. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 2006, 22, 101–127. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Rao, P.K.; Bhattacharjee, S.; Gelvin, S.B. Expression of plant protein phosphatase 2C interferes with nuclear import of the Agrobacterium T-complex protein VirD2. Proc. Natl. Acad. Sci. USA 2004, 101, 5164–5169. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Krichevsky, A.; Vaidya, M.; Tzfira, T.; Citovsky, V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. USA 2005, 102, 5733–5738. [Google Scholar] [CrossRef] [PubMed]
- Ditt, R.F.; Kerr, K.F.; de Figueiredo, P.; Delrow, J.; Comai, L.; Nester, E.W. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 2006, 19, 665–681. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N.; Spantzel, J.; Lee, L.Y.; Zhu, Y.; Lin, K.; Johnson, S.J.; Gelvin, S.B. Overexpression of several Arabidopsis histone genes increases Agrobacterium-mediated transformation and transgene expression in plants. Plant Cell 2009, 21, 3350–3367. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Vaghchhipawala, Z.; Ryu, C.M.; Kang, L.; Wang, K.; Del-Pozo, O.; Martin, G.B.; Mysore, K.S. Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol. Plant Microbe Interact. 2007, 20, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Boone, L.; Kocz, R.; Zhang, C.; Binns, A.N.; Lynn, D.G. At the maize/Agrobacterium interface: Natural factors limiting host transformation. Chem. Biol. 2000, 7, 611–621. [Google Scholar] [CrossRef]
- Joubert, P.; Beaupère, D.; Lelièvre, P.; Wadouachi, A.; Sangwan, R.S.; Sangwan-Norreel, B.S. Effects of phenolic compounds on Agrobacterium vir genes and gene transfer induction—A plausible molecular mechanism of phenol binding protein activation. Plant Sci. 2002, 162, 733–743. [Google Scholar] [CrossRef]
- Cangelosi, G.A.; Ankenbauer, R.G.; Nester, E.W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. USA 1990, 87, 6708–6712. [Google Scholar] [CrossRef] [PubMed]
- Hinchee, M.A.W.; Cnnnor-Ward, D.V.; Newell, C.A.; McDonnell, R.E.; Sato, S.J.; Gasser, C.S.; Fischhoff, D.A.; Re, D.B.; Fraley, R.T.; Horsch, R.B. Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat. Biotechnol. 1988, 6, 915–922. [Google Scholar] [CrossRef]
- Paz, M.M.; Shou, H.X.; Guo, Z.B.; Zhang, Z.Y.; Banerjee, A.K.; Wang, K. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 2004, 136, 167–179. [Google Scholar] [CrossRef]
- Dang, W.; Wei, Z.M. An optimized Agrobacterium-mediated transformation for soybean for expression of binary insect resistance genes. Plant Sci. 2007, 173, 381–389. [Google Scholar] [CrossRef]
- Liu, S.J.; Wei, Z.M.; Huang, J.Q. The effect of co-cultivation and selection parameters on Agrobacterium-mediated transformation of Chinese soybean varieties. Plant Cell Rep. 2008, 27, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, P.A.; Simmonds, D.H. Susceptibility to Agrobacterium tumefaciens and cotyledonary node transformation in short-season soybean. Plant Cell Rep. 2000, 19, 478–484. [Google Scholar] [CrossRef]
- Yah, B.; Reddy, M.S.S.; Collins, G.B. Agrobacterium tumefaciens-mediated transformation of soybean (Glycine max (L.) Merrill) using immature zygotic cotyledon explants. Plant Cell Rep. 2000, 19, 1090–1097. [Google Scholar]
- Godwin, I.; Todd, G.; Ford-Lloyd, B.; Newbury, H.J. The effects of acetosyringone and pH on Agrobacterium-mediated transformation vary according to plant species. Plant Cell Rep. 1991, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Enríquez-Obregón, G.A.; Prieto-Samsónov, D.L.; Riva, G.A.; Pérez, M.; Selman-Housein, G.; Vázquez-Padrón, R.I. Agrobacterium-mediated Japonica rice transformation: A procedure assisted by an antinecrotic treatment. Plant Cell Tissue Organ 1999, 59, 1659–168l. [Google Scholar] [CrossRef]
- Olhoft, P.M.; Somers, D.A. l-Cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledonary-node cells. Plant Cell Rep. 2001, 20, 706–710. [Google Scholar]
- Sangwan, R.S.; Bourgeois, Y.; Brown, S.; Vasseur, G.; Sangwan-Norreel, B.S. Characterization of competent cells and early events of Agrobacterium-mediated genetic transformation in Arabidopsis thaliana. Planta 1992, 188, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Chateau, S.; Sangwa, R.S.; Sangwan-Norreel, B.S. Competence of Arabidopsis thaliana genotypes and mutants for Agrobacterium tumefaciens-mediated gene transfer: Role of phytohormones. J. Exp. Bot. 2000, 51, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xu, Y. Hypocotyl-based Agrobacterium-mediated transformation of soybean (Glycine max) and application for RNA Interference. Plant Cell Rep. 2008, 27, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Sponsel, V. Gibberellins: Regulaters of plant height and seed germination adjustment factor. In Plant Physiology, 4th ed.; Taiz, L., Zeiger, E., Eds.; Academic Press: San Diego, CA, USA, 2006; p. 465. [Google Scholar]
- Karssen, C.M.; Zagorski, S.; Kepczynski, J.; Groot, S.P.C. Key role for endogenous gibberellins in the control of seed germination. Ann. Bot. 1989, 63, 71–80. [Google Scholar]
- Brits, G.J.; Cutting, J.G.M.; Brown, N.A.C.; Staden, J. Environmental and hormonal regulation of seed dormancy and germination in Cape fynbos Leucospermum R.Br. (Proteaceae) species. Plant Growth Regul. 1995, 17, 181–193. [Google Scholar] [CrossRef]
- Weyers, J.D.B.; Paterson, N.W. Plant hormones and the control of physiological processes. New Phytol. 2001, 152, 375–407. [Google Scholar] [CrossRef]
- Xue, R.G.; Zhang, B.; Xie, H.F. Overexpression of a NTR1 in transgenic soybean confers tolerance to water stress. Plant Cell Tissue Organ 2007, 89, 177–183. [Google Scholar] [CrossRef]
- Villemont, E.; Dubois, F.; Sangwan, R.S.; Vasseur, G.; Bourgeois, Y.; Sangwan-Norreel, B.S. Role of the host cell cycle in the Agrobacterium-mediated genetic transformation of Petunia: Evidence of an S-phase control mechanism for T-DNA transfer. Planta 1997, 201, 160–172. [Google Scholar] [CrossRef]
- Phillips, A.L.; Ward, D.A.; Uknes, S.; Appleford, N.E.; Lange, T.; Huttly, A.K.; Grebe, J.E.; Hedden, P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995, 108, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Riou-Khamlichi, C.; Huntley, R.; Jacqmard, A.; Murray, J.A. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 1999, 283, 1541–1544. [Google Scholar] [CrossRef] [PubMed]
- Menges, M.; de Jager, S.M.; Gruissem, W.; Murray, J.A. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005, 41, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis-structure, function, regulation. Phytochemistry 2009, 70, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.L.; Hatfield, R.D.; Samac, D.A. Cloning of an alfalfa polyphenol oxidase gene and evaluation of its potential in preventing postharvest protein degradation. J. Sci. Food Agric. 2008, 88, 1406–1414. [Google Scholar] [CrossRef]
- Rouet, M.A.; Mathieu, Y.; Barbier-Brygoo, H.; Lauriere, C. Characterization of active oxygen-producing proteins in response to hypo-osmolarity in tobacco and Arabidopsis cell suspensions: Identification of a cell wall peroxidase. J. Exp. Bot. 2006, 57, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Matsuoka, A.; Kovalchuk, I. High frequency Agrobacterium tumefaciens-mediated plant transformation induced by ammonium nitrate. Plant Cell Rep. 2009, 28, 737–757. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.G.; Babiano, M.J. Endogenous abscisic acid during the germination of chick-pea seeds. Physiol. Plant. 1997, 100, 500–504. [Google Scholar] [CrossRef]
- Crane, K.E.; Ross, C.W. Effects of wounding on cytokinin activity in cucumber cotyledons. Plant Physiol. 1986, 82, 1151–1152. [Google Scholar] [CrossRef] [PubMed]
- Haberer, G.; Kieber, J.J. Cytokinins. New insights into a classic phytohormone. Plant Physiol. 2002, 128, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 165–221. [Google Scholar] [PubMed]
- Zhang, Y.; Turner, J.G. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.A.; Lenka, S.K.; Normanly, J.; Walker, E.L.; Roberts, S.C. Methyl jasmonate represses growth and affects cell cycle progression in cultured Taxus cells. Plant Cell Rep. 2014, 33, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- Mader, J.C.; Hanke, D.E. Immunocytochemical study of cell cycle control by cytokinin in cultured soybean cells. J. Plant Growth Regul. 1996, 15, 95–102. [Google Scholar] [CrossRef]
- Murray, J.A.H.; Doonan, J.; Riou-Khamlichi, C.; Meijer, M.; Oakenfull, E.A. G1 cyclins, cytokinins and the regulation of the G1/S transition. In The Plant Cell Cycle and Its Interfaces; Francis, D., Ed.; Sheffield Academic Press: Sheffiel, UK, 2001; pp. 9–41. [Google Scholar]
- Bosch, M.; Berger, S.; Schaller, A.; Stintzi, A. Jasmonate-dependent induction of polyphenol oxidase activity in tomato foliage is important for defense against Spodoptera exigua but not against Manduca sexta. BMC Plant Biol. 2014, 14, 257. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.L.; Arruda, P.; Benedetti, C.E. An Arabidopsis gene induced by wounding functionally homologous to flavoprotein oxidoreductases. Plant Mol. Biol. 2000, 44, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Chotikacharoensuk, T.; Arteca, R.N.; Arteca, J.M. Use of differential display for the identification of touch-induced genes from an ethylene-insensitive Arabidopsis mutant and partial characterization of these genes. J. Plant Physiol. 2006, 163, 1305–1320. [Google Scholar] [CrossRef] [PubMed]
- Chehab, E.W.; Kim, S.; Savchenko, T.; Kliebenstein, D.; Dehesh, K.; Braam, J. Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol. 2011, 156, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Laukkanen, H.; Haggman, H.; Kontunen-Soppela, S.; Hohtola, A. Tissue browning of in vitro cultures of Scots pine: Role of peroxidase and polyphenol oxidase. Physiol. Plant 1999, 106, 337–343. [Google Scholar] [CrossRef]
- Wang, Y.; Li, B.; Du, M.; Eneji, A.E.; Wang, B.; Duan, L.; Li, Z.H.; Tian, X. Mechanism of phytohormone invovlement in feedback regulation of cotton leaf senescence induced by potassium deficiency. J. Exp. Bot. 2012, 63, 5887–5901. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Xu, C.N.; Wang, B.M.; Jia, J.Z. Effects of plant growth regulators on secondary wall thickening of cotton fibres. Plant Growth Regul. 2001, 35, 233–237. [Google Scholar]
- Weiler, E.W.; Jourdan, P.S.; Conrad, W. Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.R.; Singh, D. Phenolic content pattern, polyphenol oxidase and lipoxygenase activity in relation to albinism, fruit malformation and nubbins production in strawberry (Fragaria × ananassa Duch). J. Plant Biochem. Biotechonl. 2010, 19, 67–72. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Concellón, A.; Añón, M.C.; Chaves, A.R. Characterization and changes in polyphenol oxidase from eggplant fruit (Solanum melongena L.) during storage at low temperature. Food Chem. 2004, 88, 17–24. [Google Scholar] [CrossRef]
- Reuveni, R. Biochemical markers for disease resistance. In Molecular Methods in Plant Pathology; Singh, R.P., Singh, U.S., Eds.; CRC Press: Boca Raton, FL, USA, 1995; pp. 99–144. [Google Scholar]
- Brown, R.C.; Lemmon, B.E. Methods in plant immunolight microscopy. Methods Cell Biol. 1995, 49, 85–107. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Yao, X.; Zhao, M.; Zhao, Q.; Du, Y.; Yu, C.; Xie, F. Comparison of Soybean Transformation Efficiency and Plant Factors Affecting Transformation during the Agrobacterium Infection Process. Int. J. Mol. Sci. 2015, 16, 18522-18543. https://doi.org/10.3390/ijms160818522
Jia Y, Yao X, Zhao M, Zhao Q, Du Y, Yu C, Xie F. Comparison of Soybean Transformation Efficiency and Plant Factors Affecting Transformation during the Agrobacterium Infection Process. International Journal of Molecular Sciences. 2015; 16(8):18522-18543. https://doi.org/10.3390/ijms160818522
Chicago/Turabian StyleJia, Yuying, Xingdong Yao, Mingzhe Zhao, Qiang Zhao, Yanli Du, Cuimei Yu, and Futi Xie. 2015. "Comparison of Soybean Transformation Efficiency and Plant Factors Affecting Transformation during the Agrobacterium Infection Process" International Journal of Molecular Sciences 16, no. 8: 18522-18543. https://doi.org/10.3390/ijms160818522





