miR-198 Represses the Proliferation of HaCaT Cells by Targeting Cyclin D2
Abstract
:1. Introduction
2. Results and Discussion
2.1. miR-198 Represses the Proliferation of Cells
2.2. Prediction of miR-198 Binding Sites in the 3′-UTR of CCND2 mRNA

2.3. Luciferase Assay of miR-198 and CCND2 3′-UTR in HaCaT Cells

2.4. Forced Expression of MiR-198 Reduces CCND2 Expression
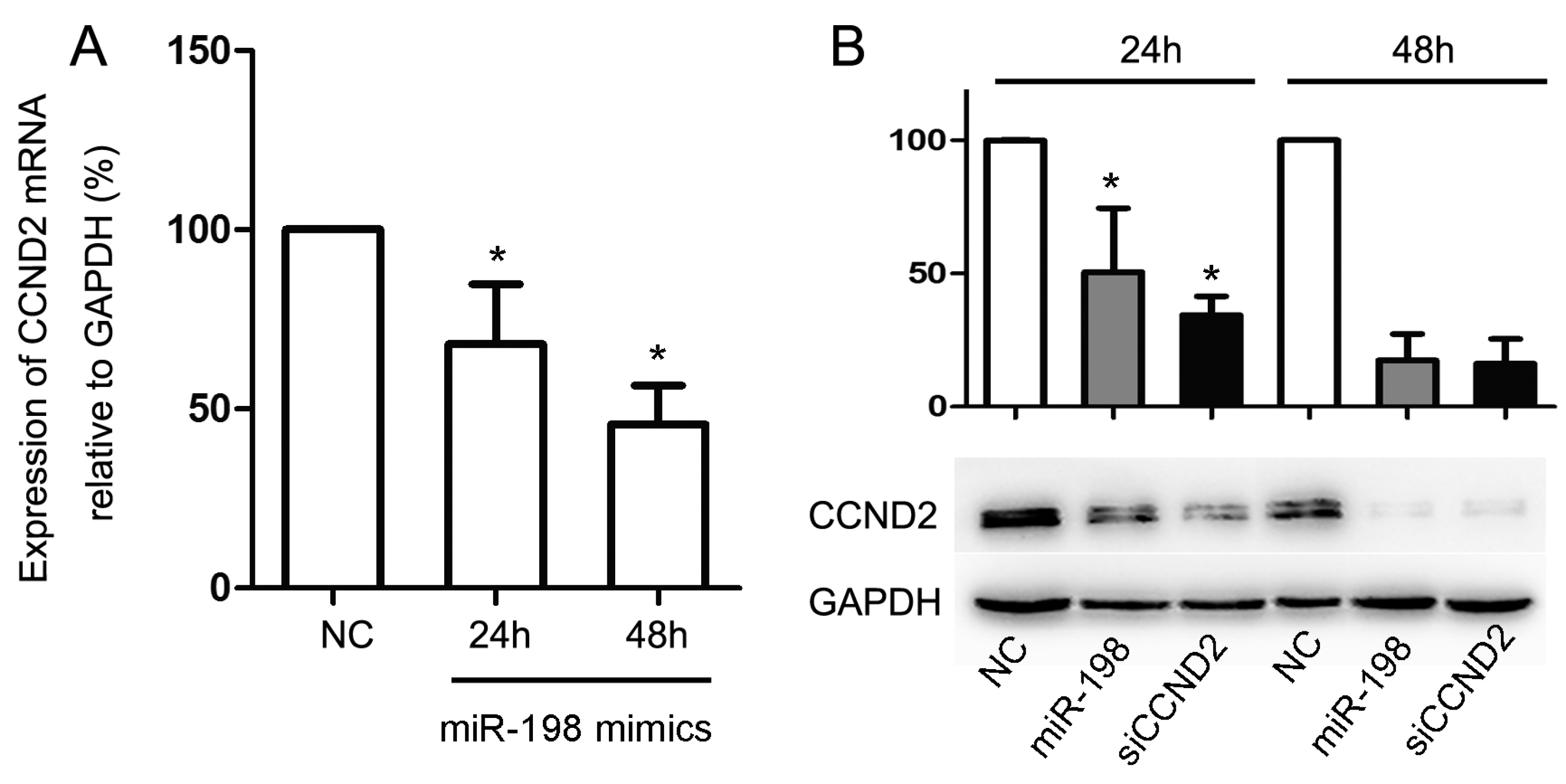
2.5. CCND2 siRNA Transfection Represses the Proliferation of Cells

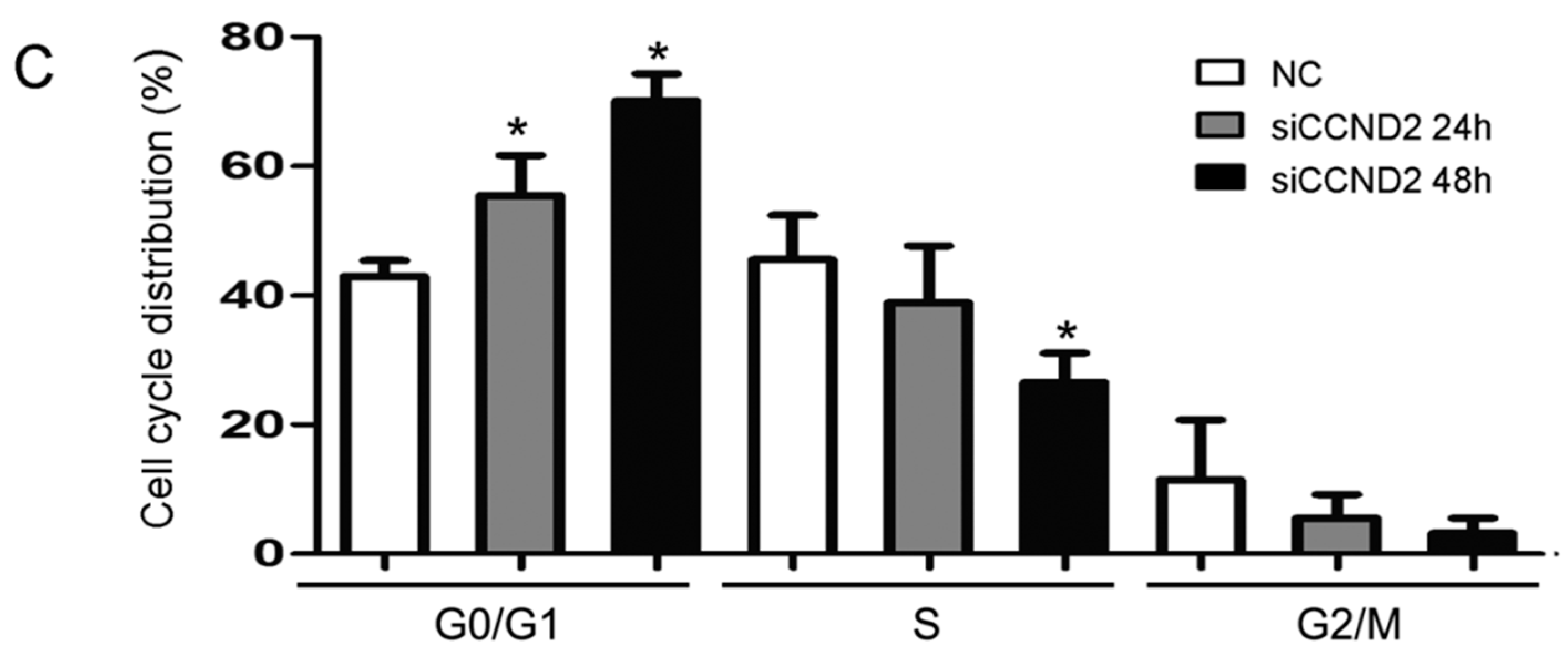
2.6. Discussion
3. Experimental Section
3.1. Cell Culture
3.2. RNA Oligonucleotides and Transfection
3.3. Quantitative Analysis of miRNAs and mRNAs
3.4. Protein Extraction and Western Blot
3.5. Luciferase Assay
3.6. Cell Proliferation
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ambros, V. The functions of animal micrornas. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding rnas in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Elfimova, N.; Noetel, A.; Varnholt, H.; Riemer, J.; Kwiecinski, M.; Quasdorff, M.J.; Dienes, H.P.; Odenthal, M. The role of tumor suppressor microRNAs in hepatocellular carcinoma. Hepatology 2009, 50. [Google Scholar] [CrossRef]
- Elfimova, N.; Sievers, E.; Eischeid, H.; Kwiecinski, M.; Noetel, A.; Hunt, H.; Becker, D.; Frommolt, P.; Quasdorff, M.; Steffen, H.M.; et al. Control of mitogenic and motogenic pathways by miR-198, diminishing hepatoma cell growth and migration. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Chonglei, B.; Zhou, J.; Chung, T.H.; Huang, G.; Yan, J.; Chng, W.J. Identification of tumor suppressive microRNAs in multiple myeloma by pharmacologic unmasking. Clin. Lymphoma Myeloma Leuk. 2013, 13, S37–S38. [Google Scholar]
- Ogunwobi, O.O.; Li, J.; Zhao, X.; Trevino, J.; George, T.; Liu, C. MiR-198 silencing is a mechanism of gemcitabine resistance in human pancreatic adenocarcinoma. Cancer Res. 2013, 73. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.; Kong, X.; Chen, H.; Wang, Y.; Qin, M.; Lin, Y.; Xu, J.; Hong, J.; Chen, Y.X.; et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Wang, J.L.; Chen, H.M.; Fang, J.Y. MicroRNA-198 regulates elf3 expression and inhibits cell proliferation in colorectal cancer. Tumor 2014, 34, 494–500. [Google Scholar]
- Yang, J.; Zhao, H.; Xin, Y.; Fan, L. MicroRNA-198 inhibits proliferation and induces apoptosis of lung cancer cells via targeting fgfr1. J. Cell. Biochem. 2014, 115, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, G.M.; Common, J.E.A.; Gopal, F.E.; Srikanta, S.; Lakshman, K.; Lunny, D.P.; Lim, T.C.; Tanavde, V.; Lane, E.B.; Sampath, P. “See-saw” expression of microRNA-198 and Fstl1 from a single transcript in wound healing. Nature 2013, 494, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Roberts, J.M. Living with or without cyclins and cyclin-dependent kinases. Gene Dev. 2004, 18, 2699–2711. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Cancer cell cycles. Science 1996, 274, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Ajchenbaum-Cymbalista, F.; Griffin, J.D. Regulation of G1/S transition by cyclins D2 and D3 in hematopoietic cells. Proc. Natl. Acad. Sci. USA 1993, 90, 9571–9575. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Bezprozvannaya, S.; Williams, A.H.; Qi, X.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008, 22, 3242–3254. [Google Scholar] [CrossRef] [PubMed]
- Kota, J.; Chivukula, R.R.; O'Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microrna delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Pandey, A.; Shukla, A.; Talwelkar, S.S.; Kumar, A.; Pant, A.B.; Parmar, D. MiR-497 and miR-302b regulate ethanol-induced neuronal cell death through bcl2 protein and cyclin D2. J. Biol. Chem. 2011, 286, 37347–37357. [Google Scholar] [CrossRef] [PubMed]
- Ekhteraei Tousi, S.; Soltani, B.; Soleimani, M.; Sadeghizadeh, M.; Mowla, S.J. Potential involvement of Hsa-miR-133b in cardiomyocytes differentiation and proliferation. Cell J. 2012, 14, 35. [Google Scholar]
- Li, L.; Sarver, A.L.; Alamgir, S.; Subramanian, S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes pax3 and ccnd2 expression in rhabdomyosarcoma. Lab. Investig. 2012, 92, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Mussnich, P.; D'Angelo, D.; Leone, V.; Croce, C.M.; Fusco, A. The high mobility group a proteins contribute to thyroid cell transformation by regulating miR-603 and miR-10b expression. Mol. Oncol. 2013, 7, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Zamore, P.D. A microRNA in a multiple-turnover rnai enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Zamore, P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002, 12, 225–232. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Dipaola, J.E.; Aspden, L.M.; Nikiforov, Y.E. MiRNA markers of aggressive behavior of papillary thyroid carcinoma. Lab. Investig. 2009, 89, 118A. [Google Scholar]
- Lulla, R.R.; Costa, F.F.; Bischof, J.M.; Chou, P.M.; Bonaldo, M.D.F.; Vanin, E.F.; Soares, M.B. Identification of differentially expressed micrornas in osteosarcoma. Sarcoma 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Li, R.; Ding, K.; Lobie, P.E.; Zhu, T. MiR-198 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the HGF/c-MET pathway. FEBS Lett. 2011, 585, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Marin-Muller, C.; Li, D.; Bharadwaj, U.; Li, M.; Chen, C.; Hodges, S.E.; Fisher, W.E.; Mo, Q.; Hung, M.C.; Yao, Q. A tumorigenic factor interactome connected through tumor suppressor microRNA-198 in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 5901–5913. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Li, S.; Ye, D.; Yang, D.; Yue, F.; Guo, Y.; Chen, X.; Chen, F.; Zhang, J.; Song, X. Livin expression may be regulated by miR-198 in human prostate cancer cell lines. Eur. J. Cancer 2013, 49, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Liu, X.B.; Wong, B.Y.H.; Ng, R.W.M.; Yuen, A.P.W.; Wei, W.I. Mature miR-184 as potential oncogenic microrna of squamous cell carcinoma of tongue. Clin.Cancer Res. 2008, 14, 2588–2592. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Yang, J.; Lin, J.; Yao, N.; Zhu, Y.; Zheng, J.; Xu, J.; Cheng, J.Q.; Lin, J.Y.; Ma, X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv. Syst. 2009, 25, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Walter, B.A.; Valera, V.A.; Pinto, P.A.; Merino, M.J. Comprehensive microrna profiling of prostate cancer. J. Cancer 2013, 4, 350–357. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Dan, G.; Shangguan, T.; Hao, H.; Tang, R.; Peng, K.; Zhao, J.; Sun, H.; Zou, Z. miR-198 Represses the Proliferation of HaCaT Cells by Targeting Cyclin D2. Int. J. Mol. Sci. 2015, 16, 17018-17028. https://doi.org/10.3390/ijms160817018
Wang J, Dan G, Shangguan T, Hao H, Tang R, Peng K, Zhao J, Sun H, Zou Z. miR-198 Represses the Proliferation of HaCaT Cells by Targeting Cyclin D2. International Journal of Molecular Sciences. 2015; 16(8):17018-17028. https://doi.org/10.3390/ijms160817018
Chicago/Turabian StyleWang, Jian, Guorong Dan, Tao Shangguan, Han Hao, Ran Tang, Kaige Peng, Jiqing Zhao, Huiqin Sun, and Zhongmin Zou. 2015. "miR-198 Represses the Proliferation of HaCaT Cells by Targeting Cyclin D2" International Journal of Molecular Sciences 16, no. 8: 17018-17028. https://doi.org/10.3390/ijms160817018




