Cloning and Characterization of Surface-Localized α-Enolase of Streptococcus iniae, an Effective Protective Antigen in Mice
Abstract
:1. Introduction
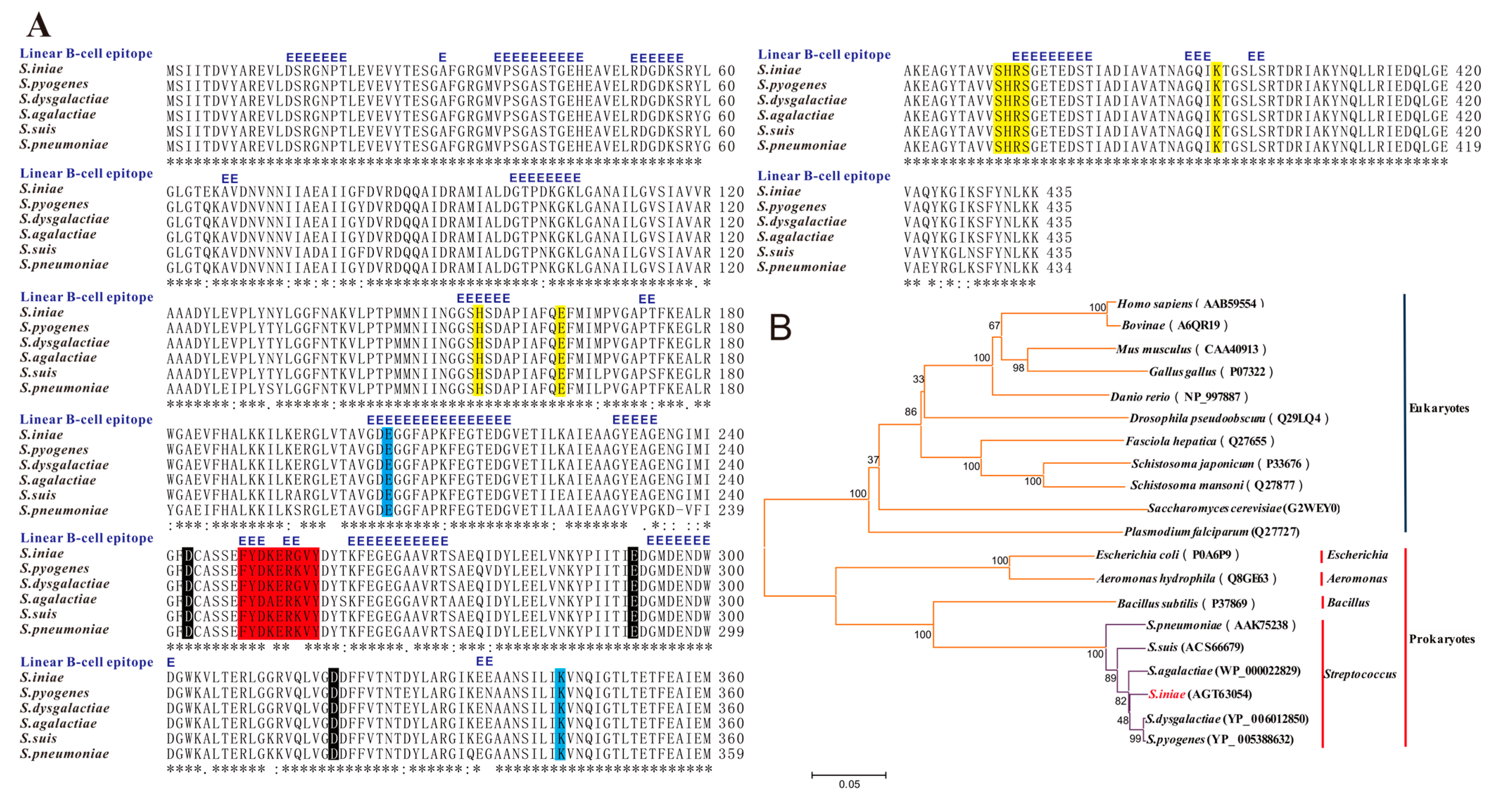
2. Results
2.1. Molecular Cloning, Expression and Characterization of S. iniae α-Enolase

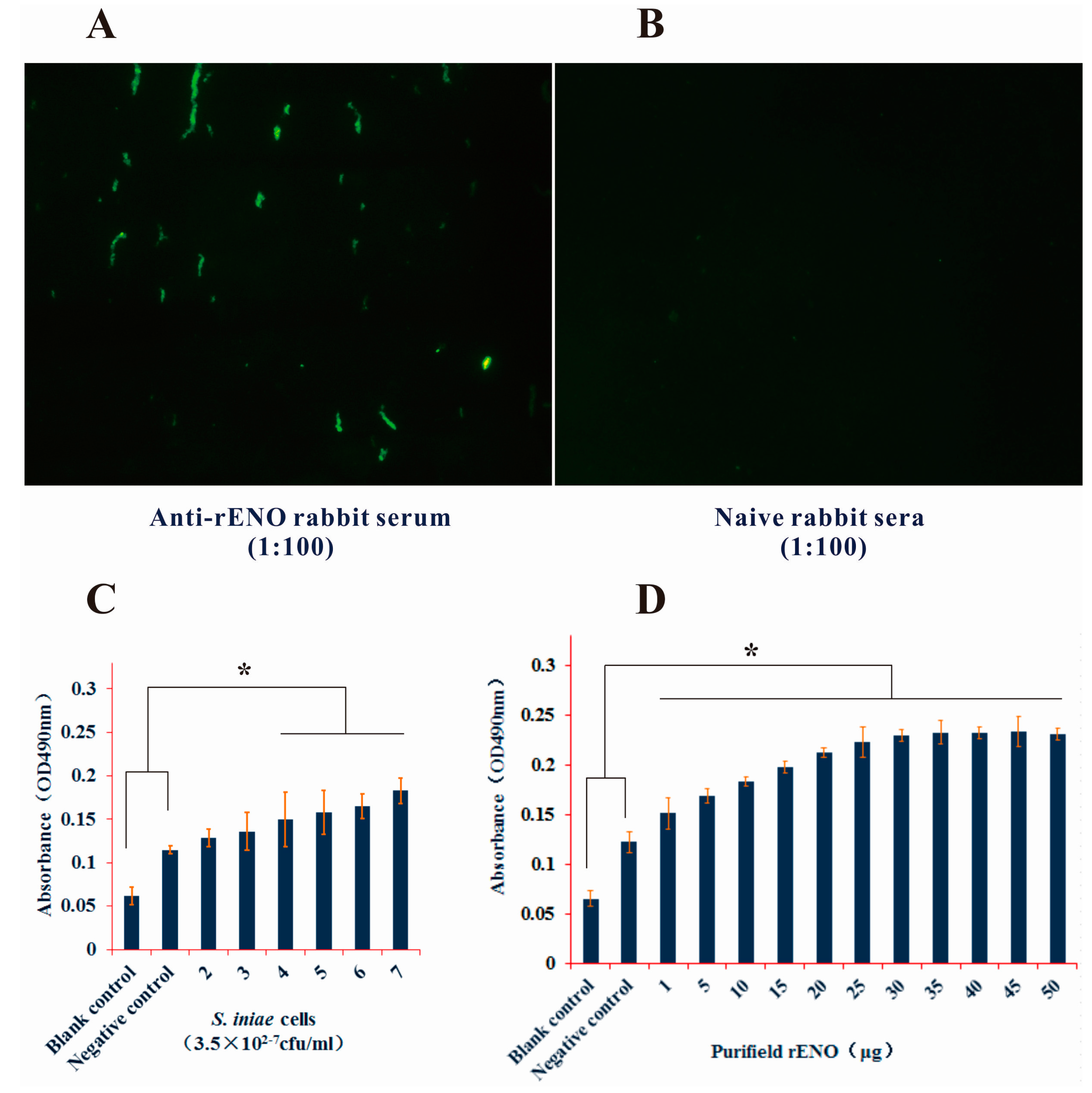
2.2. Subcellular Location of S. iniae α-Enolase
2.3. Surface Display of S. iniae α-Enolase
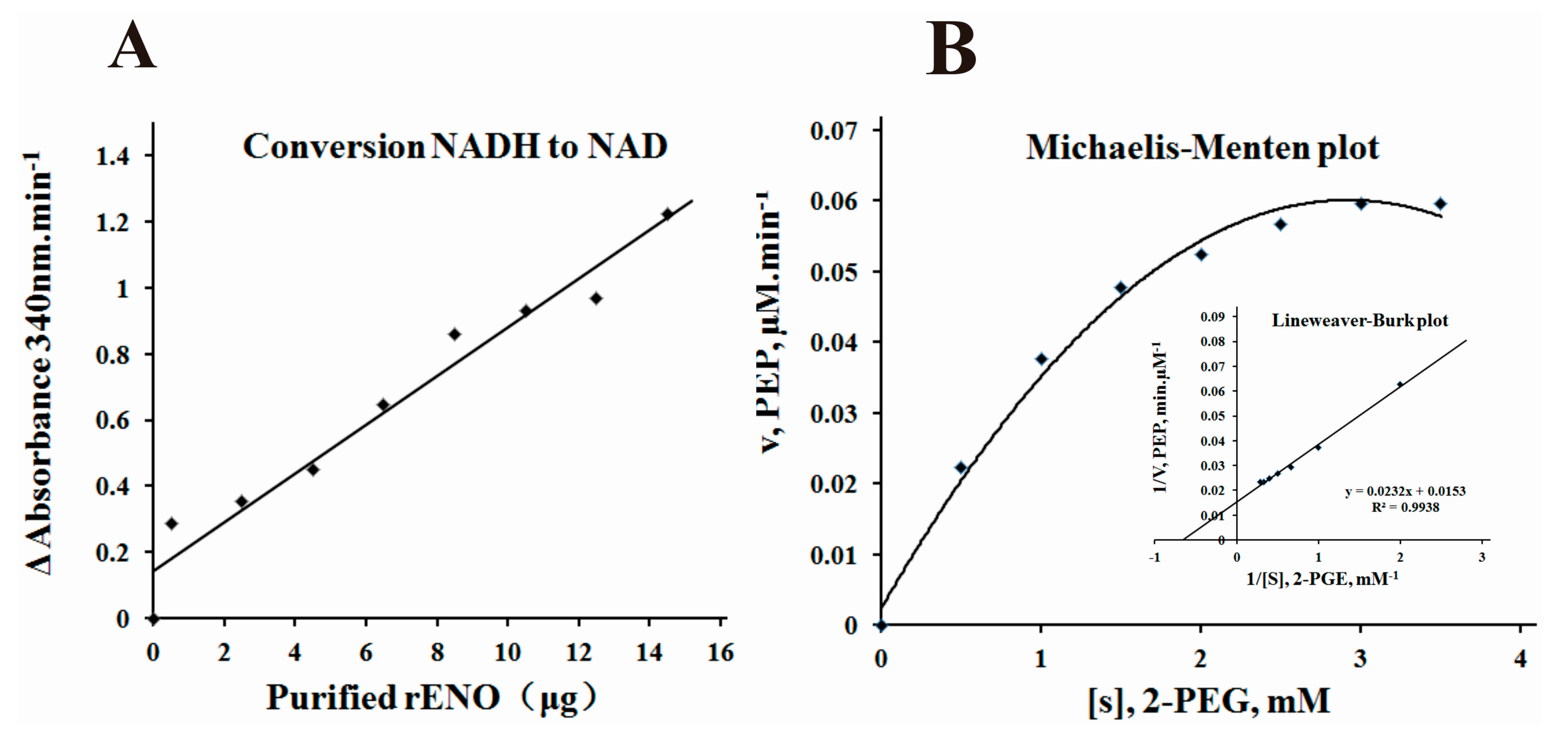
2.4. α-Enolase Activity and Enzyme Kinetics Analysis
2.5. hPlg Binding Activity of S. iniae α-Enolase

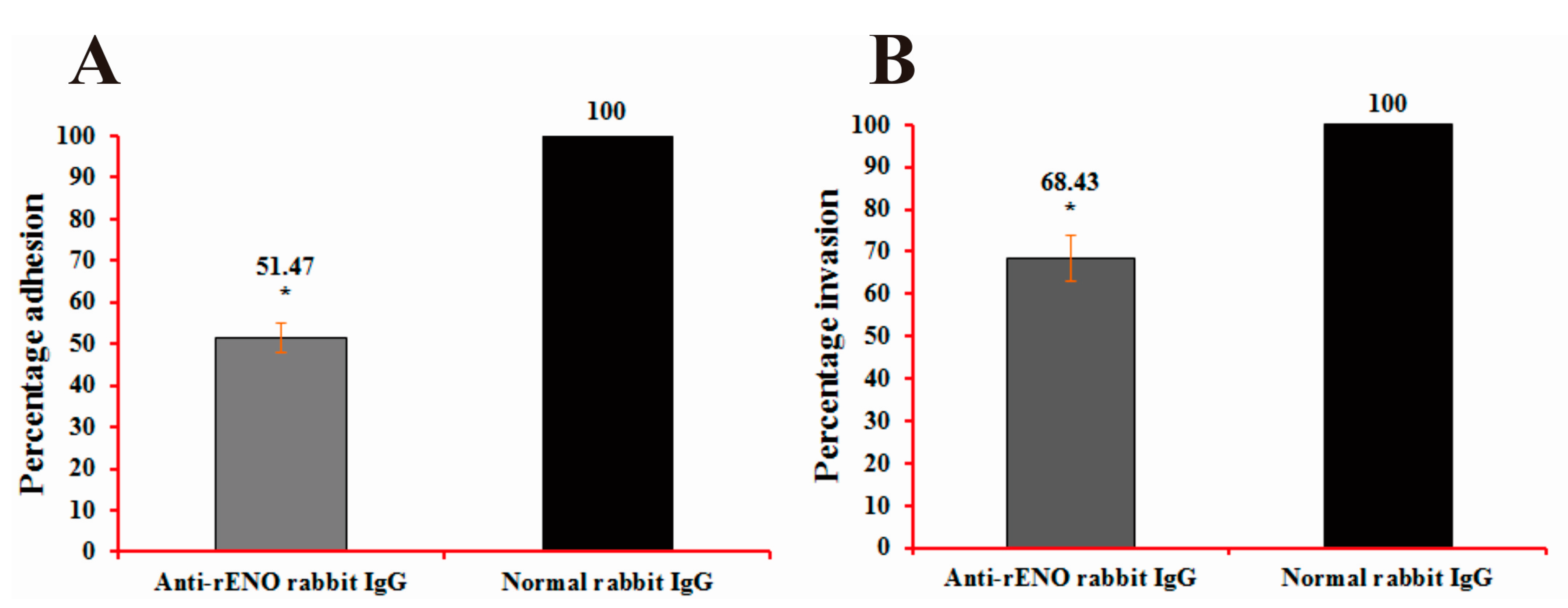
2.6. Adherence and Invasion Assays of S. iniae to BHK-21 Cells
2.7. Immunoprotection of rENO as a Subunit Vaccine against S. iniae in Mice
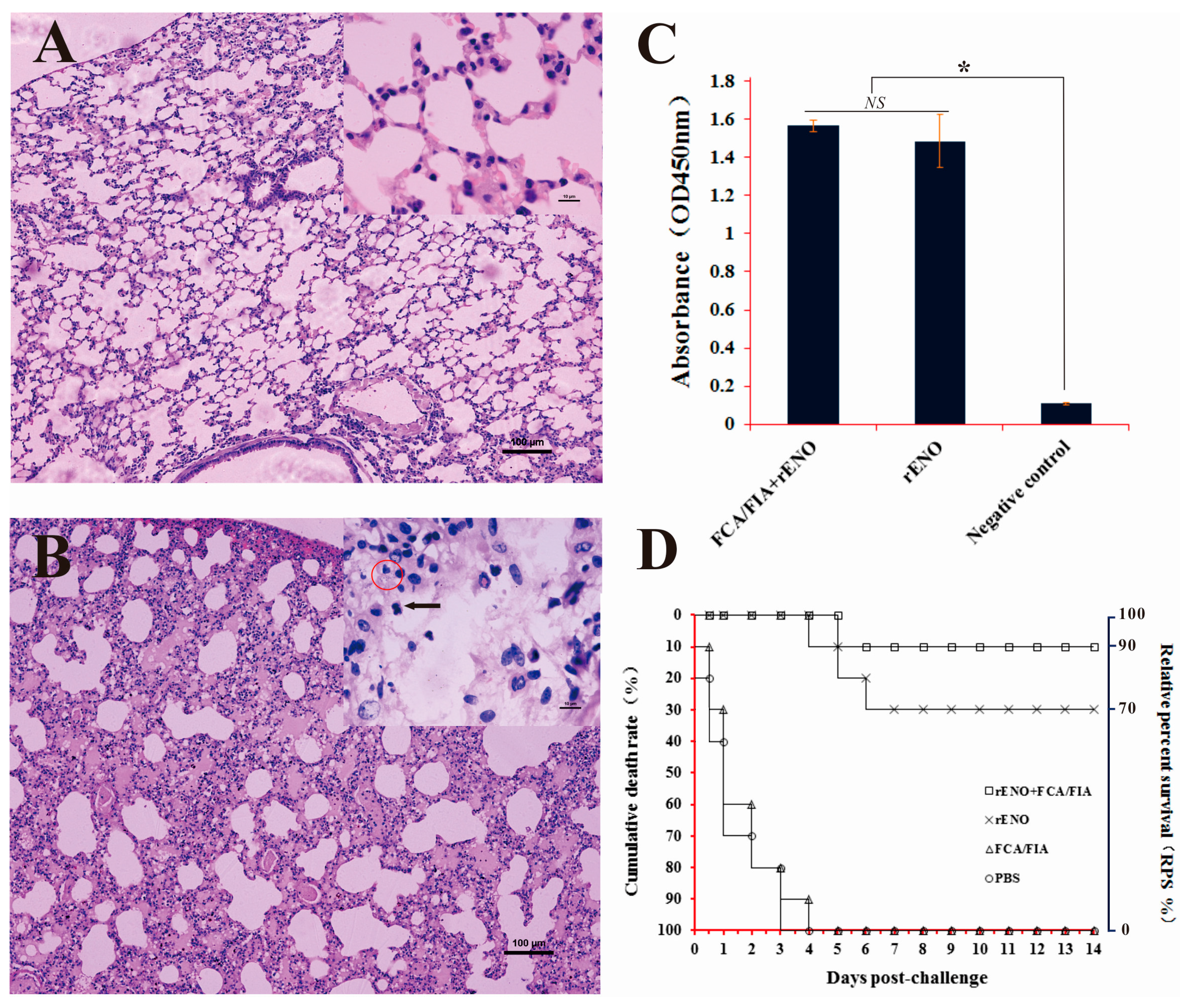
3. Discussion
4. Experimental Section
4.1. Strains, Plasmids and Media
4.2. Animals
4.3. Amplification and Bioinformatics Analyses
4.4. Cloning, Expression, and Purification of S. iniae α-Enolase
4.5. Preparation of Anti-rENO Serum
4.6. Isolation of Cellular Protein Fractions
4.7. SDS-PAGE and Western-Blot Analysis
4.8. Indirect Immunofluorescence Assay
4.9. ELISA Analysis
4.10. Determination of α-Enolase Activity
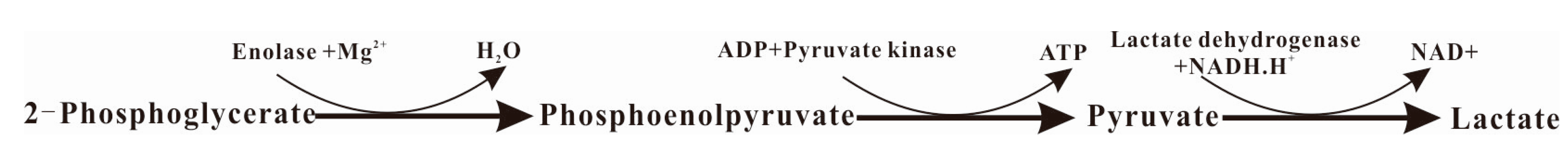
4.11. Plasminogen Interaction of α-Enolase
4.12. Adherence and Invasion Assays
4.13. Immune Protection Assay
4.14. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lau, S.K.; Woo, P.C.; Tse, H.; Leung, K.-W.; Wong, S.S.; Yuen, K.-Y. Invasive streptococcus iniae infections outside north america. J. Clin. Microbiol. 2003, 41, 1004–1009. [Google Scholar] [CrossRef] [PubMed]
- Agnew, W.; Barnes, A.C. Streptococcus iniae: An aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet. Microbiol. 2007, 122, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Facklam, R.; Elliott, J.; Shewmaker, L.; Reingold, A. Identification and characterization of sporadic isolates of streptococcus iniae isolated from humans. J. Clin. Microbiol. 2005, 43, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Koh, T.; Sng, L.H.; Yuen, S.; Thomas, C.; Tan, P.; Tan, S.; Wong, N. Streptococcal cellulitis following preparation of fresh raw seafood. Zoonoses Public Health 2009, 56, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Luk, W.-K.; Fung, A.M.; Hui, W.-T.; Fong, A.H.; Chow, C.-W.; Wong, S.S.; Yuen, K.-Y. Clinical isolates of streptococcus iniae from asia are more mucoid and β-hemolytic than those from north america. Diagn. Microbiol. Infect. Dis. 2006, 54, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-R.; Yan, J.-C.; Yeh, C.-Y.; Lee, S.-Y.; Lu, J.-J. Invasive infection with streptococcus iniae in taiwan. J. Med. Microbiol. 2007, 56, 1246–1249. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.R.; Litt, M.; Kertesz, D.A.; Wyper, P.; Rose, D.; Coulter, M.; McGeer, A.; Facklam, R.; Ostach, C.; Willey, B.M. Invasive infections due to a fish pathogen, Streptococcus iniae. N. Engl. J. Med. 1997, 337, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.N.; Pfeifer, J.D.; Caparon, M. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 2002, 70, 3904–3914. [Google Scholar] [CrossRef] [PubMed]
- Baiano, J.C.; Tumbol, R.A.; Umapathy, A.; Barnes, A.C. Identification and molecular characterisation of a fibrinogen binding protein from streptococcus iniae. BMC Microbiol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.C.; Horne, M.T.; Ellis, A.E. Streptococcus iniae expresses a cell surface non-immune trout immunoglobulin-binding factor when grown in normal trout serum. Fish Shellfish Immunol. 2003, 15, 425–431. [Google Scholar] [CrossRef]
- Locke, J.B.; Colvin, K.M.; Datta, A.K.; Patel, S.K.; Naidu, N.N.; Neely, M.N.; Nizet, V.; Buchanan, J.T. Streptococcus iniae capsule impairs phagocytic clearance and contributes to virulence in fish. J. Bacteriol. 2007, 189, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Rohde, M.; Polok, A.; Preissner, K.T.; Chhatwal, G.S.; Bergmann, S. Cooperative plasminogen recruitment to the surface of streptococcus canis via m protein and enolase enhances bacterial survival. MBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, S.V.; Smith, A.A.; Qin, J.-H.; Pal, U. A surface enolase participates in Borrelia burgdorferi-plasminogen interaction and contributes to pathogen survival within feeding ticks. Infect. Immun. 2012, 80, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.; Madureira, P.; Andrade, E.B.; Bouaboud, A.; Morello, E.; Ferreira, P.; Poyart, C.; Trieu-Cuot, P.; Dramsi, S. Group B streptococcus gapdh is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLoS ONE 2012, 7, e29963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledo, A.; Coleman, J.; Kuhlow, C.; Crowley, J.; Benach, J. The enolase of Borrelia burgdorferi is a plasminogen receptor released in outer membrane vesicles. Infect. Immun. 2012, 80, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Han, X.; Wang, Q.; Li, X.; Yi, L.; Liu, Y.; Ding, C. Vibrio parahaemolyticus enolase is an adhesion-related factor that binds plasminogen and functions as a protective antigen. Appl. Microbiol. Biotechnol. 2014, 98, 4937–4948. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V. Multifunctional α-enolase: Its role in diseases. Cell. Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Kulshreshtha, P.; Bambah Mukku, D.; Bhatnagar, R. α-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta Proteins Proteomics 2008, 1784, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Floden, A.M.; Watt, J.A.; Brissette, C.A. Borrelia burgdorferi enolase is a surface-exposed plasminogen binding protein. PLoS ONE 2011, 6, e27502. [Google Scholar] [CrossRef] [PubMed]
- Marcos, C.M.; Fátima da Silva, J.; Oliveira, H.C.; Moraes da Silva, R.A.; Mendes-Giannini, M.J.S.; Fusco-Almeida, A.M. Surface-expressed enolase contributes to the adhesion of Paracoccidioides brasiliensis to host cells. FEMS Yeast Res. 2012, 12, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Pan, X.; Sun, W.; Wang, C.; Zhang, H.; Li, X.; Ma, Y.; Shao, Z.; Ge, J.; Zheng, F. Streptococcus suis enolase functions as a protective antigen displayed on the bacterial cell surface. J. Infect. Dis. 2009, 200, 1583–1592. [Google Scholar] [CrossRef] [PubMed]
- Dinis, M.; Tavares, D.; Veiga-Malta, I.; Fonseca, A.J.; Andrade, E.B.; Trigo, G.; Ribeiro, A.; Videira, A.; Cabrita, A.M.S.; Ferreira, P. Oral therapeutic vaccination with Streptococcus sobrinus recombinant enolase confers protection against dental caries in rats. J. Infect. Dis. 2009, 199, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, J.; Aase, A.; Bergmann, S.; Herstad, T.K.; Rødal, G.; Frank, R.; Rohde, M.; Hammerschmidt, S. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology 2006, 152, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Esgleas, M.; Li, Y.; Hancock, M.A.; Harel, J.; Dubreuil, J.D.; Gottschalk, M. Isolation and characterization of α-enolase, a novel fibronectin-binding protein from streptococcus suis. Microbiology 2008, 154, 2668–2679. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, C.J.; Radin, J.N.; Sublett, J.E.; Gao, G.; Kaushal, D.; Tuomanen, E.I. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 2004, 72, 5582–5596. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V.; Fischetti, V.A. Α-enolase, a novel strong plasmin (ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 1998, 273, 14503–14515. [Google Scholar] [CrossRef] [PubMed]
- Pridgeon, J.W.; Klesius, P.H. Major bacterial diseases in aquaculture and their vaccine development. Anim. Sci. Rev. 2013. [Google Scholar] [CrossRef]
- Baiano, J.C.; Barnes, A.C. Towards control of Streptococcus iniae. Emerg. Infect. Dis. 2009, 15, 1891. [Google Scholar] [CrossRef] [PubMed]
- Chen, D. Study on Etiology, Pathology and Diagnostic Method of the Streptococcus Iniae Infection in Channel Catfish (Ictalurus Punctatus). Ph.D. Thesis, Sichuan Agriculture University, Ya’an, China, 2011. [Google Scholar]
- Cole, J.N.; Ramirez, R.D.; Currie, B.J.; Cordwell, S.J.; Djordjevic, S.P.; Walker, M.J. Surface analyses and immune reactivities of major cell wall-associated proteins of group A streptococcus. Infect. Immun. 2005, 73, 3137–3146. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Schoenen, H.; Hammerschmidt, S. The interaction between bacterial enolase and plasminogen promotes adherence of Streptococcus pneumoniae to epithelial and endothelial cells. Int. J. Med. Microbiol. 2013, 303, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Rohde, M.; Chhatwal, G.S.; Hammerschmidt, S. A-enolase of Streptococcus pneumoniae is a plasmin (ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 2001, 40, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Berge, A.; Sjöbring, U. Pam, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 1993, 268, 25417–25424. [Google Scholar] [PubMed]
- Pancholi, V.; Fischetti, V.A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 1992, 176, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Wild, D.; Diekmann, O.; Frank, R.; Bracht, D.; Chhatwal, G.S.; Hammerschmidt, S. Identification of a novel plasmin (ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Mol. Microbiol. 2003, 49, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Plow, E.; Herren, T.; Redlitz, A.; Miles, L.; Hoover-Plow, J. The cell biology of the plasminogen system. FASEB J. 1995, 9, 939–945. [Google Scholar] [PubMed]
- Jones, M.N.; Holt, R.G. Cloning and characterization of an α-enolase of the oral pathogen Streptococcus mutans that binds human plasminogen. Biochem. Biophys. Res. Commun. 2007, 364, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.L.; Sellati, T.J.; Testa, J.E.; Kew, R.R.; Furie, M.B.; Benach, J.L. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 1995, 63, 2478–2484. [Google Scholar] [PubMed]
- Zhang, X.; Ge, Y.; Zhao, S.; Hu, Y.; Ashman, R.B. Immunisation with the glycolytic enzyme enolase confers effective protection against Candida albicans infection in mice. Vaccine 2011, 29, 5526–5533. [Google Scholar]
- Chen, N.; Yuan, Z.-G.; Xu, M.-J.; Zhou, D.-H.; Zhang, X.-X.; Zhang, Y.-Z.; Wang, X.-W.; Yan, C.; Lin, R.-Q.; Zhu, X.-Q. Ascaris suum enolase is a potential vaccine candidate against ascariasis. Vaccine 2012, 30, 3478–3482. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Zhao, G.; Xu, H.; Wu, W.; Du, W.; Huang, J.; Yu, X.; Hu, X. Reverse vaccinology approach identify an echinococcus granulosus tegumental membrane protein enolase as vaccine candidate. Parasitol. Res. 2010, 106, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, W.; Tian, Y.; Mao, Q.; Lv, X.; Shang, M.; Li, X.; Yu, X.; Huang, Y. Surface display of Clonorchis sinensis enolase on Bacillus subtilis spores potentializes an oral vaccine candidate. Vaccine 2014, 32, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- LaFrentz, B.R.; Shoemaker, C.A.; Klesius, P.H. Immunoproteomic analysis of the antibody response obtained in nile tilapia following vaccination with a Streptococcus iniae vaccine. Vet. Microbiol. 2011, 152, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-C.; Zhang, J.; Sun, L. Streptococcus iniae sf1: Complete genome sequence, proteomic profile, and immunoprotective antigens. PLoS ONE 2014, 9, e91324. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-F.; Wang, K.-Y.; Geng, Y.; Wang, J.; Huang, L.-Y.; Li, J.-M. Streptococcus iniae isolated from channel catfish (Ictalurus punctatus) in China. Available online: http://cmsadmin.atp.co.il/Content_siamb/editor/63.2011.593.Wang,%207%20pages.pdf (accessed on 17 June 2015).
- Lowe, B.A.; Miller, J.D.; Neely, M.N. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect. Immun. 2007, 75, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, S.; Yan, Y.; Zhang, Z.; Li, D.; Yu, H.; Wang, C.; Nong, X.; Zhou, X.; Gu, X. Potential of recombinant inorganic pyrophosphatase antigen as a new vaccine candidate against Baylisascaris schroederi in mice. Vet. Res. 2013, 44, 90. [Google Scholar] [CrossRef] [PubMed]
- Hüther, F.; Psarros, N.; Duschner, H. Isolation, characterization, and inhibition kinetics of enolase from Streptococcus rattus FA-1. Infect. Immun. 1990, 58, 1043–1047. [Google Scholar] [PubMed]
- Zhang, M.; Sun, K.; Sun, L. Regulation of autoinducer 2 production and luxs expression in a pathogenic Edwardsiella tarda strain. Microbiology 2008, 154, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, C.-S.; Sun, L. Comparative study of the immune effect of an Edwardsiella tarda antigen in two forms: Subunit vaccine vs DNA vaccine. Vaccine 2011, 29, 2051–2057. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Wang, K.; Chen, D.; Geng, Y.; Huang, X.; He, Y.; Ji, L.; Liu, T.; Wang, E.; Yang, Q.; et al. Cloning and Characterization of Surface-Localized α-Enolase of Streptococcus iniae, an Effective Protective Antigen in Mice. Int. J. Mol. Sci. 2015, 16, 14490-14510. https://doi.org/10.3390/ijms160714490
Wang J, Wang K, Chen D, Geng Y, Huang X, He Y, Ji L, Liu T, Wang E, Yang Q, et al. Cloning and Characterization of Surface-Localized α-Enolase of Streptococcus iniae, an Effective Protective Antigen in Mice. International Journal of Molecular Sciences. 2015; 16(7):14490-14510. https://doi.org/10.3390/ijms160714490
Chicago/Turabian StyleWang, Jun, Kaiyu Wang, Defang Chen, Yi Geng, Xiaoli Huang, Yang He, Lili Ji, Tao Liu, Erlong Wang, Qian Yang, and et al. 2015. "Cloning and Characterization of Surface-Localized α-Enolase of Streptococcus iniae, an Effective Protective Antigen in Mice" International Journal of Molecular Sciences 16, no. 7: 14490-14510. https://doi.org/10.3390/ijms160714490




